November 28, 2008
Facial anthropometry of northern Iran
The effect of ethnicity on facial anthropometry in Northern Iran.
Jahanshahi M, Golalipour MJ, Heidari K.
Introduction: The evaluation and measurement of human body dimensions are achieved by physical anthropometry. Cephalometery is a branch of anthropometry science in which the head and face anatomical dimensions are measured. This research was conducted in view of the importance of anthropometric indices of the face in forensic medicine, surgery, paediatrics and medical imaging. Methods: This descriptive and cross-sectional study was set up to determine and compare the face shapes in Fars and Turkman ethnic groups of 808 normal 17- to 20-year-old males and females in Gorgon, North Iran (Fars group 407, male 200 and female 207; Turkman group 401, male 198 and female 203). The length and width of faces were determined by using classic cephalometery technique with Martin spreading callipers, and the shape of faces in the ethnic group of Fars and Turkman in both sexes was compared. Results: The dominant type of face shape in both the native Fars and Turkman females was euryprosopic (37.7 and 51.7 percent, respectively). The dominant type of face shape in the native Fars and Turkman males was mesoprosopic (44 and 38.4 percent, respectively). Conclusion: This study determined the possible effect of ethnicity on the diversity of face shapes in young males and females in this region.
Link
November 27, 2008
Korean-Japanese anthropometric differences

We are able to show that average Korean head and face shapes differ significantly from Japanese head and face shapes. Koreans have higher BMI than Japanese, and Korean males and females have the same BMI, but the BMI has higher coefficients for males than for females in Japanese.
...
Japanese have a longer head and a larger head length. Nose breadth and mouth breadth of Japanese are larger than those for Koreans, and Japanese have a wider distance between the pupils of the eyes.
...Only ORT, OLT and BMI for Korean male subjects were larger than those for Japanese male subjects; the other Japanese items (weight, TH, RTV, LTV, REV, LEV, SVH, MFH, HL, ORE, OLE, OS, HB, BB, BA, NH, NB, MB, and ID) were higher than those for Koreans. The MB values, in particular, showed the largest difference between Korean and Japanese male subjects.
In the comparison of Korean and Japanese female head and face dimensions, almost all item values show statistically significant differences between the Korean [End Page 325] and Japanese groups, except in the categories of height, HC, ORE, and OLE. HC did not show statistically significant differences between the two female subject groups. Weight, BMI, OS, ORT, OLT, BB, HC, SA, and BIGB for Korean females were larger than those values for Japanese females.
Human Biology Volume 80, Number 3
Comparison of Korean and Japanese Head and Face Anthropometric Characteristics
Hyun-Ja Lee, Se-Jin Park
Abstract
The primary purpose of this research is to collate, compare, and discuss the presently available data for head and face dimensions among Korean and Japanese ethnic groups. Classification of Korean male and female head and face types is simpler than classification of Japanese subjects. Male groups have more statistically significant morphological differences than females, and Japanese subjects display larger values for head and face measurement categories than Korean subjects. Japanese item values for head and face dimensions show distinct differences between male and female subjects compared to Korean subjects. Japanese male subjects have distinct differences from Korean male subjects and relatively lower values for head and face dimensions as age increases compared to Korean male subjects. Generally, female subjects have no regular tendency according to age compared to male subjects.
Link
mtDNA of Ancient Koreans
Genetic Characterization and Assessment of Authenticity of Ancient Korean Skeletal Remains
Hwan Young Lee et al.
Abstract
To study the maternal lineage history of Korea, we extracted DNA from the skeletal remains of 35 museum samples (some dating back to the Paleolithic Age) excavated from 11 local burial sites scattered throughout southern Korea. Mitochondrial DNA (mtDNA) control region sequences (HV1, HV2, and HV3) were successfully determined for 11 samples with no sharing of the control region polymorphisms with individuals involved in the laboratory analyses. Each of the 11 mtDNAs was assigned to the appropriate East Asian mtDNA haplogroup according to the haplogroup-specific control region mutation motif and diagnostic coding region single nucleotide polymorphism. The successful mtDNA haplogroup determination for each ancient Korean mtDNA and the confirmation of the absence of abnormal mutations based on the haplogroup-directed database comparisons indicates that there is no mosaic structure from cross-contamination or sample mix-up or other errors in our mtDNA sequences. The presence of haplogroups B, D, and G in the prehistoric age is consistent with the hypothesis that the early Korean population has a common origin in the northern regions of the Altai Mountains and Lake Baikal of southeastern Siberia. In addition, the modern Korean population, which possesses lineages from both southern and northern haplogroups, suggests additional gene flow from southern Asian haplogroups in recent times, but many more ancient samples need to be analyzed to directly tell whether there was regional continuity or replacement of early lineages by other lineages in ancient Korea.
Link
mtDNA of Old Believers
Russian Old Believers: Genetic Consequences of Their Persecution and Exile, as Shown by Mitochondrial DNA Evidence
Samara Rubinstein et al.
Abstract
In 1653, the Patriarch Nikon modified liturgical practices to bring the Russian Orthodox Church in line with those of the Eastern (Greek) Orthodox Church, from which it had split 200 years earlier. The Old Believers (staroveri) rejected these changes and continued to worship using the earlier practices. These actions resulted in their persecution by the Russian Orthodox Church, which forced them into exile across Siberia. Given their history, we investigate whether populations of Old Believers have diverged genetically from other Slavic populations as a result of their isolation. We also examine whether the three Old Believer populations analyzed in this study are part of a single gene pool (founder population) or are instead derived from heterogeneous sources. As part of this analysis, we survey the mitochondrial DNAs (mtDNAs) of 189 Russian Old Believer individuals from three populations in Siberia and 201 ethnic Russians from different parts of Siberia for phylogenetically informative mutations in the coding and noncoding regions. Our results indicate that the Old Believers have not significantly diverged genetically from other Slavic populations over the 200–300 years of their isolation in Siberia. However, they do show some unique patterns of mtDNA variation relative to other Slavic groups, such as a high frequency of subhaplogroup U4, a surprisingly low frequency of haplogroup H, and low frequencies of the rare East Eurasian subhaplogroup D5.
Link
November 26, 2008
Article on Greek citizenship and naturalization
Greece, like most European countries, does not give automatic citizenship to children born in the country. And becoming a naturalized citizen is a long, difficult process: Greece makes it harder than almost any other country in the European Union – only Austria is tougher. It’s also the only one of the original EU 15 that makes no special provisions for children born to immigrants in the country. Members of the Greek diaspora, in contrast, can get a passport easily.While much can be done to improve the existing legal framework, I am fundamentally in agreement with the basic principle of jus sanguinis, citizenship by descent, rather than jus soli, citizenship by birthplace.
How should one become a Greek citizen? This is an elaboration and amendment of my earlier thoughts on this matter:
- The offspring of a Greek citizen are Greek citizens.
- Non-citizens may be naturalized by Law passed with increased Majority of 2/3 of the Parliament. (General provisions should be made for persons of Greek origin, although individual persons of non-Greek origin who rendered extraordinary service to the Nation should also be allowed to become citizens).
- Spouses of Greek citizens become permanent residents, and may become Greek citizens after ten years of marriage and demonstrable knowledge of Greek language and culture. They cannot, however, confer their citizenship to offspring from non-Greek mates (as in #1)
- Legal immigrants have renewable residence permits (for work, study, etc.) of specific time length.
- Legal immigrants may obtain permanent residency (of unspecified time length) after 10 years of immigrant status and demonstrable knowledge of Greek language and culture.
- Children found in Greece become Greek citizens; if they are claimed by their non-Greek parents before the age of 18, then the provisions of #7 apply to them.
- Children educated in Greece are allowed in the country for the duration of their studies, and become permanent residents thereafter.
- Illegal immigrants may not be legalized within the country; they may apply for legal immigrant status after departing from Greece.
I believe that such a system is flexible enough to facilitate honest legal immigrants in their everyday life. On the other hand, it also ensures that the Greek citizen body will always consist of ethnic Greeks and their descendants.
Allele surfing vs. Positive Selection
From the paper:
The survey of the HGDP database on human polymorphisms reveals that large allele frequency differences between continental regions are extremely common. Indeed as much as 30% of loci show very large allele frequency differences between continents. These differences are unlikely to have been created by positive selection, but are more likely the result of neutral demographic processes such as the surfing phenomenon. Because the erosion of large allele frequency differences by mutation is slow, even for large mutation rates, the surprisingly large number of strongly differentiated STR alleles also do not need to be explained by the action of positive selection. Africa and the Americas show a much larger extent of differentiation than Eurasia or East Asia, which is certainly due to changes in allele frequencies during the colonisation of the Eurasian and the American continents. Disentangling the effects of selection and neutral demographic processes on genome diversity remains an important challenge of future human evolution studies.
This is a serious challenge to the selectionist paradigm and should be answered by its proponents. I would say that, from now on, the "gold standard" of positive selection should be concrete evidence that the proposed selected alleles actually do something that could have been selected, e.g., lactase persistence, where allele frequency differences are combined with a specific trait, which in turn is correlated with a particular selective influence (milk consumption after weaning). Statistical inference of selection without a comprehensive explanation is no longer intellectually convincing.
For example, ASPM and MCPH1 are loci that generated a lot of excitement as selection targets due to their large inter-group frequency differences. However, followup work has not found any substantial associations between them and anything of value: Has ASPM been the target of recent selection?, ASPM, MCPH1, CDK5RAP and BRCA1 and general cognition, reading or language. Were they really selected, or did they ride the wave of human advance?
Annals of Human Genetics doi: 10.1111/j.1469-1809.2008.00489.x
Large Allele Frequency Differences between Human Continental Groups are more Likely to have Occurred by Drift During range Expansions than by Selection
T. Hofer et al.
Several studies have found strikingly different allele frequencies between continents. This has been mainly interpreted as being due to local adaptation. However, demographic factors can generate similar patterns. Namely, allelic surfing during a population range expansion may increase the frequency of alleles in newly colonised areas. In this study, we examined 772 STRs, 210 diallelic indels, and 2834 SNPs typed in 53 human populations worldwide under the HGDP-CEPH Diversity Panel to determine to which extent allele frequency differs among four regions (Africa, Eurasia, East Asia, and America). We find that large allele frequency differences between continents are surprisingly common, and that Africa and America show the largest number of loci with extreme frequency differences. Moreover, more STR alleles have increased rather than decreased in frequency outside Africa, as expected under allelic surfing. Finally, there is no relationship between the extent of allele frequency differences and proximity to genes, as would be expected under selection. We therefore conclude that most of the observed large allele frequency differences between continents result from demography rather than from positive selection.
Link
November 25, 2008
Neolithic Expansion was not gradual
The expansion of the agricultural system on the map, as represented by the earlier Neolithic, was not uniform and regular across Europe as a whole, but proceeded in leaps. Using a scale of 500-year isochrones, several leaps are identifiable: at 8000 crossing the Taurus barrier, 6700-6100 crossing the southern Adriatic barrier, 6100-5600 crossing the Central European agro-ecological barrier and 5000-4000 BC expanding on the other, marginal zones. Using a vector map build-up, 10 points of renewed expansion and 9 accretion or contact zones were detected. Clearly, the whole does not correspond to a process of homogeneous diffusion, approximately steady, but a process marked by phases of geographical expansion and stasis, already noted by Guilaine (2000, 2003) and Guilaine and Manen (2007) for the LBK and the Mediterranean axis, with their so-called ”arrhythmic" model, which can be generalized across the whole of Europe.Journal of Archaeological Science doi: 10.1016/j.jas.2008.11.004
Detection of diffusion and contact zones of early farming in europe
from the space-time distribution of 14c dates
Jean-Pierre Bocquet-Appel et al.
Abstract
The spread of early farming in Europe is revisited using a sample of 3072 audited 14C CalBC dates from 940 georeferenced early Neolithic sites. The surface expansion of early Neolithic has been reconstituted using the kriging technique of spatial interpolation. Centres of renewed expansion, of contact zones, and the main routes of expansion have been highlighted by means of a vector map, representing the gradient. The expansion of the agricultural system on the map, was not uniform and regular across Europe as a whole, but proceeded in leaps. With the scale of detection of the 500-year isochrones, several leaps are identifiable: at 8000 calBC crossing the Taurus barrier, 6700-6100 calBC crossing the southern Adriatic barrier, 6100-5600 calBC crossing the Central European agro-ecological barrier and 5000-4000 calBC expanding on the other, marginal zones. Using a vector map, 10 points of renewed expansion and 9 contact zones, were detected. The whole does not correspond to a process of homogeneous diffusion, approximately steady, but a process marked by phases of geographical expansion and stasis.
Link
Origin of ancient Chinese horses from ancient DNA
To shed light on the origin of the diverse haplogroups, we studied the distribution of the seven haplogroups in 17 modern populations, and showed that only F and D have a significant geographical pattern (Table 3 and Fig.2). Haplogroup F is prevalent in East Eurasian populations and its frequencies declines from east to west, consistent with the results of previous studies (McGahern et al., 2006). Based on our findings in Chinese horses older than 4000 years, we propose that haplogroup F is an ancient haplogroup of East Asian origin.
...
we suggest that the origin of Chinese domestic horses is more complex than previously thought, and that both indigenous breeds and introduced maternal lineages were involved in the process of domestication. Thus, our data fail to support either of the two previous hypotheses on the domestication of horses in China.
...
Phylogenetic analysis revealed that the Przewalski fell into cluster A2, distinct
from Chinese ancient horses and modern horses. Genetic distance analysis indicated that the Przewalski were far from the ancient horses, suggesting that the Przewalski horse and the domestic horse should be considered sister taxa with a common ancestor, and that the Przewalski horse is not the wild ancestor to the Chinese domestic horse. However, this conclusion should be viewed with caution, as the Przewalski horse underwent a severe genetic bottleneck: all present individuals descend from only 13 survivors, with only four maternal lineages (Volf & Kus, 1991).
Related: Wikipedia article on Przewalski's horse. Also, Lippold & Hofreiter from ISBA3:
We sequence 600 bp of mtDNA from the mitochondrial D-Loop of different individuals obtained from different locations. Our initial results indicate tremendous genetic diversity, but no phylogeographic pattern within this marker. The genetic distribution of these ancient samples also falls within the broad diversity range apparent in recent horses.It would be interesting to see more ancient DNA results for wild and domestic horses, especially from the Eurasiatic steppes where they are believed to have undergone domestication.
Journal of Archaeological Science doi: 10.1016/j.jas.2008.11.006
Ancient DNA provides new insights into the origin of the Chinese
domestic horse
Dawei Cai et al.
Abstract
Domestic horses played a pivotal role in ancient China, but their exact origin remains controversial. To investigate the origin of Chinese domestic horses, we analyzed mitochondrial DNA (mtDNA) from 35 horse remains, aged between 4000 and 2000 years, excavated from nine archaeological sites in northern China. The Chinese ancient horses exhibited high matrilineal diversity, falling into all the seven haplogroups (A-G) observed in modern horses. These results suggest that several maternal lines were introduced into the gene pool of Chinese horses in the past. Haplogroup A and F were more prevalent in ancient horses than the other haplogroups. Interestingly, only haplogroups A and F were present in the samples older than 4000 years, while the more recent horses (between 2000 and 3000 years BP) fell into all seven haplogroups. Comparison with DNA data of present-day horses suggests haplogroup F is like to be an ancient haplogroup of East Asian origin. These analyses also suggest that the origin of Chinese domestic horses is complex, and external mtDNA input occurred after initial domestication. Our results indicate that the Chinese ancient horses are more related to the modern Mongolian horses. Lastly, our results cannot support cannot support the previous hypothesis that early Chinese domestic horses were derived from the Przewalski horse.
Link
November 23, 2008
Palatine torus in Anatolia
Why did the frequency of palatine torus increase in the ancient Anatolian populations?
S. Eroğluand Y.S. Erdal
Abstract
The frequency of the palatine torus varies in various populations from different regions of the world. In this study, the change of frequency of palatine torus is examined using 387 skulls from 12 different ancient Anatolian populations in various periods ranging from the Early Bronze Age to the first quarter of the 20th century.
While the frequency of palatine torus is 45% in the Early Bronze Age, this ratio steadily increases to 87% in the Ottoman Period and finally declines to 40% during the recent period. It was determined that the increase in the frequency of the palatine torus is statistically significant across different periods from the Early Bronze Age until the 20th century. Constituting a passageway between Asia and Europe and being located on the crossroads of the most important trade route of the Medieval Period, the Silk Road, Anatolia is known to have been subjected to recurring invasions and migrations since the 11th century A.D. Therefore, it is possible to say that, with the introduction of the Mongoloid influence in Anatolia, genetic flow has had a significant role in the observed increase in the frequency of this trait.
Link
November 21, 2008
European population structure with 300K SNPs and 6,000 individuals
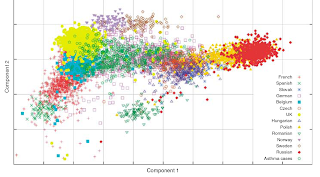 This is a nice paper on European population structure with a large number of SNPs, which comes at the heels of some earlier studies:
This is a nice paper on European population structure with a large number of SNPs, which comes at the heels of some earlier studies:- Genetic structure in Northern Europe with 250K SNPs
- Geography and Genetic structure in Europe (again)
- 500K SNP Europe-wide study of genetic structure
- In this study the first principal component of variation is along an east-west axis, rather than north-south as in previous studies. This is due to the limited number of southern European populations, and the great number of populations along an east-west axis from Spain to Russia. As I have mentioned before, the results of a principal components analysis are dataset-dependent.
- The nice technique of this paper is to infer the ancestry of an unknown sample (which could be perhaps a forensic case or customer of an ancestry analysis test) using only summary statistics. Imagine that you have 1,000 individuals from different populations, and want to guess the ancestry of an unknown test case. You could go about doing a full STRUCTURE run using the 1,001 individuals, or you could exploit the information garnered from an analysis of the 1,000 individuals (a PCA analysis in this case) to test the 1,001th individual. This is much faster and convenient, since the full STRUCTURE run is very time consuming.
Some ethnic groups are clearly distinguishable from each other (e.g. Swedes vs. Spaniards); some groups are partitioned into fairly disjoint sets (Spain I vs. Catalans in Spain II); others mutually overlap (e.g., British and Irish); while others overlap asymetrically (e.g., some former Yugoslavs in the Greek cluster, but not vice versa).In this paper, the authors did a systematic study of the "ethnic distinctiveness" of their samples. In a first experiment, they used 80% of their data to identify the features of the various populations (e.g., Germans or Spaniards), and then tried to guess the origin of the remaining 20%:
 It is clear that some nations appear to be distinct. For example, most test Spaniards (94.5%) are correctly guessed as Spaniards, with some (5.5%) guessed as French. Of course, this distinctiveness would be reduced if further populations (e.g. the Portuguese) were added to the analysis. More strongly, 99.1% of Norwegians are guessed correctly as Norwegians.
It is clear that some nations appear to be distinct. For example, most test Spaniards (94.5%) are correctly guessed as Spaniards, with some (5.5%) guessed as French. Of course, this distinctiveness would be reduced if further populations (e.g. the Portuguese) were added to the analysis. More strongly, 99.1% of Norwegians are guessed correctly as Norwegians.Other nations appear to be less distinct. For example, only 45.3% of Slovaks are guessed as Slovaks with most of the remaining ones guessed as Czechs (25%) or Hungarians (22%).
In some cases there is asymmetry of affiliation. For example, no Belgians are guessed as Germans but 10.2% of Germans are guessed as Belgians. Similarly 9.9% of Swedes are guessed as Norwegians, but only 1% of Norwegians are guessed as Swedes. While each case needs to be addressed individually, this observation is consistent with historical asymmetry in immigration patterns or ethnic identity formation. So, while e.g., the bulk of Germans (64.4%) are guessed correctly, sizeable minorities are guessed as Czechs, Belgians, or Scandinavians.
I would speculate that large central European countries have historically (both due to prestige or geographical position) absorbed more diverse populations from neighboring nations, while smaller peripheral countries have mostly acted as sources of population, reserving their own genetic distinctiveness.
In a second experiment the authors guessed the origin of individuals, but excluding the country from which they actually originated.
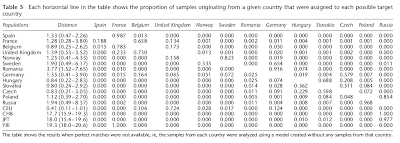 Once again, it is clear that members of particular nations can mostly be mistaken for members of their closest neighbors. Almost all Spaniards are guessed as French; French mostly as Belgians but with sizable Spanish and UK minorities; UK as Belgians but with sizable French minorities; Norwegians mostly as Swedes but with some UK; Swedes mainly as Germans but many as Norwegians; most Poles as Russians, but some Slovaks or Czechs, and so on.
Once again, it is clear that members of particular nations can mostly be mistaken for members of their closest neighbors. Almost all Spaniards are guessed as French; French mostly as Belgians but with sizable Spanish and UK minorities; UK as Belgians but with sizable French minorities; Norwegians mostly as Swedes but with some UK; Swedes mainly as Germans but many as Norwegians; most Poles as Russians, but some Slovaks or Czechs, and so on.The importance of these results can't be underestimated. While it can be argued that some ethnic groups are spuriously distinctive only due to insufficient sampling of the geographical continuum, it is more difficult to do this for others. For example, it is now possible to identify particular ethnic groups, e.g., Norwegians, with great accuracy from DNA.
More markers and more populations will doubtlessly enhance our ability to distinguish European nations using DNA. But perfect accuracy is unlikely; in most European nations there are probably minorities which -for historical reasons- allied themselves with one country or political entity even though they were ultimately of different genetic background than the majority population of that entity.
Nonetheless, at a time when -due to a sort of mental hysteresis- proclamations that "races are social constructs" are still routinely made, the discovery that not only races, but even closely related ethnic groups (e.g. Norwegians and Swedes) can be distinguished with greater than 90% accuracy, serves to illustrate the scientific irrelevance of the ethnic nihilists and the affirmation that nations are, at least in part, genetic entities.
European Journal of Human Genetics (2008) 16, 1413–1429; doi:10.1038/ejhg.2008.210
Investigation of the fine structure of European populations with applications to disease association studies
Simon C Heath et al.
Abstract
An investigation into fine-scale European population structure was carried out using high-density genetic variation on nearly 6000 individuals originating from across Europe. The individuals were collected as control samples and were genotyped with more than 300 000 SNPs in genome-wide association studies using the Illumina Infinium platform. A major East–West gradient from Russian (Moscow) samples to Spanish samples was identified as the first principal component (PC) of the genetic diversity. The second PC identified a North–South gradient from Norway and Sweden to Romania and Spain. Variation of frequencies at markers in three separate genomic regions, surrounding LCT, HLA and HERC2, were strongly associated with this gradient. The next 18 PCs also accounted for a significant proportion of genetic diversity observed in the sample. We present a method to predict the ethnic origin of samples by comparing the sample genotypes with those from a reference set of samples of known origin. These predictions can be performed using just summary information on the known samples, and individual genotype data are not required. We discuss issues raised by these data and analyses for association studies including the matching of case-only cohorts to appropriate pre-collected control samples for genome-wide association studies.
Link
Y chromosomes from Algeria
8 E3a-M2 (or 8.6% Sub-Saharan contribution)
6 E3b1-M78
40 E3b2-M81
5 J2f-M67
1 R1-M173
11 R1b3-M269
1 Q-M242
The presence of R1-M173 seems interesting, with the haplotype: 16 14 24 30 16 15 13,16 13 10 12 23 14 12 14 12 18, over (DYS456 DYS389I DYS390 DYS389II DYS458 DYS19 DYS385 DYS393 DYS391 DYS439 DYS635 DYS392 YGATAH4 DYS437 DYS438 DYS448). Does this look like an R1b (but not R-M269)? Could it be related to the unexpected R1* found in Cameroon?
Int J Legal Med. 2008 May;122(3):251-5. Epub 2007 Oct 2.
Analysis of Y-chromosomal SNP haplogroups and STR haplotypes in an Algerian population sample.
Robino C, Crobu F, Di Gaetano C, Bekada A, Benhamamouch S, Cerutti N, Piazza A, Inturri S, Torre C.
The distribution of Y-chromosomal single nucleotide polymorphism (SNP) haplogroups and short tandem repeat (STR) haplotypes was determined in a sample of 102 unrelated men of Arab origin from northwestern Algeria (Oran area). A total of nine different haplogroups were identified by a panel of 22 binary markers. The most common haplogroups observed in the Algerian population were E3b2 (45.1%) and J1 (22.5%). Y-STR typing by a 17-loci multiplex system allowed 93 haplotypes to be defined (88 were unique). Striking differences in the allele distribution and gene diversity of Y-STR markers between haplogroups could be found. In particular, intermediate alleles at locus DYS458 specifically characterized the haplotypes of individuals carrying haplogroup J1. All the intermediate alleles shared a common repeat sequence structure, supporting the hypothesis that the variant originated from a single mutational event.
Link
November 19, 2008
Y-chromosomes and mtDNA from Eulau (Corded Ware Culture, Germany)
Three male individuals, a 40-60 year-old male and two young boys (4-5 and 8-9 years old) from grave 99 belonged to Y-haplogroup R1a. The boys were related to their parent:
Additionally, the Y chromosome haplogroup R1a of the boys corresponds with the man’s (ind. 3). It is noteworthy that in grave 99 the orientation of the adult individuals follows the established pattern of the CWC, whereas both children clearly deviate from it. It appears that the burial orientation pattern was overruled for each boy to face a parent to express a biological relationship.The results have been added in my compendium of ancient Y-chromosome studies.
The mtDNA results included haplogroups:
K1b (three from grave 99) 16093C, 16224C, 16311C, 16319A
U5b (one from grave 99) 16189C, 16192T, 16270T
I (one from grave 90) 16129A, 16223T, 16391A
H (one from grave 98) 16093C, 16221T
X2 (two from grave 98) 16189C, 16223T, 16278T 73G, 153G, 195C, 225A, 226C, 263G
K1a2 (one from grave 93) 16145A, 16224C, 16311C
The occurrence of X2 and K1b suggests changes in frequency to the present-day:
The detection of mtDNA haplotypes X2 and K1b, both being very rare in modern-day European populations, is unlikely to be based on the occurrence ofFor the X2 sequence:
independent contamination events.
At present, three exact matches were found among individuals from Iran, Syria, and Estonia showing HVS I and HVS II patterns identical with the two Eulau individuals, but differing from each other by further coding region polymorphisms.Regarding the present-day distribution of X2:
Overall, it appears that the populations of the Near East, the Caucasus, and Mediterranean Europe harbor subhaplogroup X2 at higher frequencies than those of northern and northeastern Europe (P less than .05) and that X2 is rare in Eastern European as well as Central Asian, Siberian, and Indian populations and is virtually absent in the Finno-Ugric and Turkic-speaking people of the Volga-Ural region.For the K1b sequences:
Today, haplogroup K has a frequency of around 6% in Europe (6, 39). The identical sequence haplotype of individuals 1, 2, and 4 has previously been observed (4, 5).The haplogroup I is also interesting, since it was found at a high frequency in Vikings and Iron Age Danes but has a lower frequency in modern times.
...
So far this haplotype has uniquely been reported in two modern Shugnans of Tadzhikistan (5)
So, in general, these results once again point towards a change in the mtDNA gene pool across the millennia, explained by the authors:
However, we are aware of the fact that the modern distribution might not reflect the haplotype distribution during the Late Neolithic. The mitochondrial haplogroup distribution of a population is likely to have changed throughout time by factors like genetic drift or events like migration or genetic palimpsest (40).Study (41) also by Haak et al. was about Linearbandkeramik farmers from Central Europe. Study (42) by Sampietro et al. was about Neolithic Iberians.
As we recently showed, the frequencies of haplogroups could have been significantly different between Neolithic and present populations (41), although a similar study (from a different region in Europe) provides evidence of genetic continuity throughout the millennia (42).
PNAS doi: 10.1073/pnas.0807592105
Ancient DNA, Strontium isotopes, and osteological analyses shed light on social and kinship organization of the Later Stone Age
Wolfgang Haak et al.
Abstract
In 2005 four outstanding multiple burials were discovered near Eulau, Germany. The 4,600-year-old graves contained groups of adults and children buried facing each other. Skeletal and artifactual evidence and the simultaneous interment of the individuals suggest the supposed families fell victim to a violent event. In a multidisciplinary approach, archaeological, anthropological, geochemical (radiogenic isotopes), and molecular genetic (ancient DNA) methods were applied to these unique burials. Using autosomal, mitochondrial, and Y-chromosomal markers, we identified genetic kinship among the individuals. A direct child-parent relationship was detected in one burial, providing the oldest molecular genetic evidence of a nuclear family. Strontium isotope analyses point to different origins for males and children versus females. By this approach, we gain insight into a Late Stone Age society, which appears to have been exogamous and patrilocal, and in which genetic kinship seems to be a focal point of social organization.
Link
November 18, 2008
More on Henn et al. on time-dependent evolutionary mutation rate
To date, the timing of most population events in human evolutionary genetics was estimated has used a rate close to the slower one we see for older lineages, before the end of the Ice Age. So our understanding of the genetic history of early human evolution shouldn’t change very much. But the timing of the splits between mitochondrial lineages associated with relatively recent events, such as agricultural expansions, may need revision. Using our newly calibrated mitochondrial mutation rates, researchers will be better able to correlate genetic, archaeological and linguistic data, leading to a more accurate understanding of human prehistory.Thus, event ages have been systematically underestimated in the literature whenever a slower "evolutionary" mutation rate, derived from calibration has been used.
For example, according to the recent paper on Soqotra:
For calculation of time to most recent common ancestor (TMRCA), q statistics (mean divergence from inferred ancestral haplotype) were used with a HVS-I mutation rate of one transition per 20,180 years (Forster et al., 1996).Forster et al. (1996):
Therefore, we use ρ as an appropriate measure for time depth (table 2), yielding a rate of one transition per 20,180 + 1,000 years in the mtDNA control region (np 16090-16365).Thus, this rate is equivalent to 0.28/bp/My (assuming a generation length of 20 years).
But from Henn et al.:
The average of ρ-based rate estimates for events younger than 5,000 years is 0.895/bp/My, virtually identical to the pedigree-based average and well within the confidence interval calculated by Howell et al. (2003).Hence, the 6,000-year old estimate for the novel lineages identified by Cerny et al. for Soqotra should be downgraded to around ~2,000 years, making them more likely to be quite recent founder lineages, rather than Holocene lineages associated with the early settlement of the island.
Heterozygote Instability and Population-size dependent autosomal microsatellite mutation rate
Instead of population size interacting with a fixed mutation rate to determine an equilibrium heterozygosity, our analysis suggests that a feedback loop can operate causing heterozygosity to increase over time, each increase also increasing the mutation rate which in turn raises heterozygosity. Within this framework, changes in population size may act to modulate the rate of change of heterozygosity, with population expansion causing acceleration and population contraction causing a slowdown. For example, population decline erodes genetic variability through drift, reducing heterozygosity and hence potentially lowering the genome-wide mutation rate relative to a related population that has not declined.Microsatellite mutations seem to occur more often if the two alleles (we're talking about autosomal microsatellites) differ in length. Thus, heterozygosity affects the mutation rate. Populations differ in their heterozygosity, hence their microsatellites evolve at different rates. Moreover, these rates fluctuate with population size. In a growing population, heterozygosity increases, while small populations tend to be more homozygous. Since mutation rates are used to infer the ages of population splits and other events, this has important implications about human history, although these are not exactly spelled out in the paper.
BMC Genetics 2008, 9:72doi:10.1186/1471-2156-9-72
Heterozygosity increases microsatellite mutation rate, linking it to demographic historyWilliam Amos et al.
Abstract (provisional)
Background
Biochemical experiments in yeast suggest a possible mechanism that would cause heterozygous sites to mutate faster than equivalent homozygous sites. If such a process operates, it could undermine a key assumption at the core of population genetic theory, namely that mutation rate and population size are indpendent, because population expansion would increase heterozygosity that in turn would increase mutation rate. Here we test this hypothesis using both direct counting of microsatellite mutations in human pedigrees and an analysis of the relationship between microsatellite length and patterns of demographically-induced variation in heterozygosity.
ResultsWe find that microsatellite alleles of any given length are more likely to mutate when their homologue is unusually different in length. Furthermore, microsatellite lengths in human populations do not vary randomly, but instead exhibit highly predictable trends with both distance from Africa, a surrogate measure of genome-wide heterozygosity, and modern population size. This predictability remains even after statistically controlling for non-independence due to shared ancestry among populations.
ConclusionsOur results reveal patterns that are unexpected under classical population genetic theory, where no mechanism exists capable of linking allele length to extrinsic variables such as geography or population size. However, the predictability of microsatellite length is consistent with heterozygote instability and suggest that this has an important impact on microsatellite evolution. Whether similar processes impact on single nucleotide polymorphisms remains unclear.
November 16, 2008
Y chromosomes and mtDNA from Soqotra
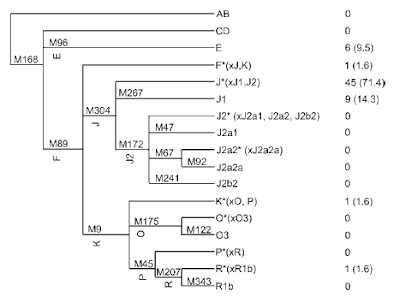 UPDATE (18/11): see a related post on the age of the new mtDNA lineages [end update]
UPDATE (18/11): see a related post on the age of the new mtDNA lineages [end update]Soqotra is an island in the Indian Ocean which belongs to Yemen. What is most interesting -to me- about this paper, is that 71.4% of the Y-chromosomes belong to haplogroup J*(xJ1, J2) which is found at trace frequencies elsewhere. Interestingly, haplogroup J2 is not found in this isolated region, while haplogroup J1 is found at a frequency of 14.3%.
The authors write:
For the perspective of the Y-chromosome data, a highThe early dates from Cadenas et al. are due to the use of an evolutionary mutation rate, and thus need to be downgraded to about ~1,000BC onwards, coinciding (within wide confidence intervals) with the formation of the earliest Arabian kingdoms. It seems probable that J*(xJ1, J2) was commoner in the past, and contributed to the population of Soqotra, but this population was later overwhelmed by the expansion of J1-carriers who dominate the Arabian peninsula to this day.
frequency of haplogroup J1 in Soqotra is consistent with
a gradient of this haplogroup in the Arabian Peninsula
(Cadenas et al., 2008). These authors estimated ages for
J1 in Arabia (9.7 +/ 2.4 in Yemen, 7.4 +/- 2.3 in Qatar and
6.4 +/- 1.4 KYBP in UAE), consistent with a Neolithic
expansion from the north (where Y-chromosome STR diversity
is higher). However, we report a much higher frequency
of J* (lack of M267 and M172) in Soqotra. Since
this lineage was not found by Cadenas et al. (2008) in
the Arabian Peninsula, this raises the possibility of an
earlier input for these lineages or more probably very
strong genetic drift of a low frequency Arabian lineage
in the Y-chromosome gene pool of Soqotra.
It is unfortunate that apparently many J2 downstream markers were typed even though there is no J2 on the island, whereas haplogroup E, occurring at a frequency of 9.5% was not further resolved. This underscores the need for a more flexible typing stratgy; at this level it is not clear whether this E came to the island from Africa or from Arabia.
American Journal of Physical Anthropology doi: 10.1002/ajpa.20960
Out of Arabia - The settlement of Island Soqotra as revealed by mitochondrial and Y chromosome genetic diversity
Viktor Cerny et al.
Abstract
The Soqotra archipelago is one of the most isolated landmasses in the world, situated at the mouth of the Gulf of Aden between the Horn of Africa and southern Arabia. The main island of Soqotra lies not far from the proposed southern migration route of anatomically modern humans out of Africa 60,000 years ago (kya), suggesting the island may harbor traces of that first dispersal. Nothing is known about the timing and origin of the first Soqotri settlers. The oldest historical visitors to the island in the 15th century reported only the presence of an ancient population. We collected samples throughout the island and analyzed mitochondrial DNA and Y-chromosomal variation. We found little African influence among the indigenous people of the island. Although the island population likely experienced founder effects, links to the Arabian Peninsula or southwestern Asia can still be found. In comparison with datasets from neighboring regions, the Soqotri population shows evidence of long-term isolation and autochthonous evolution of several mitochondrial haplogroups. Specifically, we identified two high-frequency founder lineages that have not been detected in any other populations and classified them as a new R0a1a1 subclade. Recent expansion of the novel lineages is consistent with a Holocene settlement of the island ~6 kya.
Link
November 14, 2008
mtDNA haplogroups and AIDS progression
AIDS. 2008 Nov 30;22(18):2429-2439.
Mitochondrial DNA haplogroups influence AIDS progression.
Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, Poole JC, Lautenberger J, Sezgin E, Kingsley L, Goedert JJ, Vlahov D, Donfield S, Wallace DC, Oʼbrien SJ.
OBJECTIVE:: Mitochondrial function plays a role in both AIDS progression and HAART toxicity; therefore, we sought to determine whether mitochondrial DNA variation revealed novel AIDS restriction genes, particularly as mitochondrial DNA single-nucleotide polymorphisms are known to influence regulation of oxidative phosphorylation, reactive oxygen species production, and apoptosis. DESIGN:: This is a retrospective cohort study. METHODS:: We performed an association study of mitochondrial DNA haplogroups among 1833 European American HIV-1 patients from five US cohorts: the Multicenter AIDS Cohort Study, the San Francisco City Clinic Study, Hemophilia Growth and Development Study, the Multicenter Hemophilia Cohort Study, and the AIDS Linked to Intravenous Experiences cohort to determine whether the mitochondrial DNA haplogroup correlated with AIDS progression rate. RESULTS:: Mitochondrial DNA haplogroups J and U5a were elevated among HIV-1 infected people who display accelerated progression to AIDS and death. Haplogroups Uk, H3, and IWX appeared to be highly protective against AIDS progression. CONCLUSION:: The associations found in our study appear to support a functional explanation by which mitochondrial DNA variation among haplogroups, influencing ATP production, reactive oxygen species generation, and apoptosis, is correlated to AIDS disease progression; however, repeating these results in cohorts with different ethnic backgrounds would be informative. These data suggest that mitochondrial genes are important indicators of AIDS disease progression in HIV-1 infected persons.
Link
mtDNA and goat domestication
 Wikiedia artiles on goat and bezoar.
Wikiedia artiles on goat and bezoar.The lowly goat (aiga) has played a great role in Greek history: the Aegean Sea (from Aegeus, the father of Theseus) is named after it. So were Aigai, the ancient Macedonian capital coinciding with the archaeological site of Vergina. The ancestor of the Dorians was King Aegimius, whose mythological story of contact with Hercules parallels the historical relationship between the Dorians and their Heraclid rulers. The Aegos Potami (goat rivers) is one of many aig- sites, famous among other things as the landing site of a meteor, according to the Parian Marble.
And, of course, the he-goat (tragos), associated with the god Dionysus, played its role in the Greek invention of theater, and in particular tragedy by Thespis.
Proc Natl Acad Sci U S A. 2008 Nov 12. [Epub ahead of print]
The goat domestication process inferred from large-scale mitochondrial DNA analysis of wild and domestic individuals.
Naderi S, Rezaei HR, Pompanon F, Blum MG, Negrini R, Naghash HR, Balkiz O, Mashkour M, Gaggiotti OE, Ajmone-Marsan P, Kence A, Vigne JD, Taberlet P.
The emergence of farming during the Neolithic transition, including the domestication of livestock, was a critical point in the evolution of human kind. The goat (Capra hircus) was one of the first domesticated ungulates. In this study, we compared the genetic diversity of domestic goats to that of the modern representatives of their wild ancestor, the bezoar, by analyzing 473 samples collected over the whole distribution range of the latter species. This partly confirms and significantly clarifies the goat domestication scenario already proposed by archaeological evidence. All of the mitochondrial DNA haplogroups found in current domestic goats have also been found in the bezoar. The geographic distribution of these haplogroups in the wild ancestor allowed the localization of the main domestication centers. We found no haplotype that could have been domesticated in the eastern half of the Iranian Plateau, nor further to the east. A signature of population expansion in bezoars of the C haplogroup suggests an early domestication center on the Central Iranian Plateau (Yazd and Kerman Provinces) and in the Southern Zagros (Fars Province), possibly corresponding to the management of wild flocks. However, the contribution of this center to the current domestic goat population is rather low (1.4%). We also found a second domestication center covering a large area in Eastern Anatolia, and possibly in Northern and Central Zagros. This last domestication center is the likely origin of almost all domestic goats today. This finding is consistent with archaeological data identifying Eastern Anatolia as an important domestication center.
Link
Tribute to W. W. Howells
I have previously used this dataset in some earlier posts (search "Howells") and in my article on Model-Based Clustering of World Craniometric Variation (also as pdf), in which I repeated his clustering of world human populations, but using a computationally expensive model-based clustering algorithm (conceptually similar to STRUCTURE in genetics) that was not available during his time.
One of his lesser achievements was being the mentor at Harvard of the recently deceased author Michael Crichton.
American Journal of Physical Anthropology
Volume 137 Issue S47, Pages 118 - 126
From the shoulders of a giant: Perspectives on the legacy of William White Howells (1908-2005)
Laurie R. Godfrey
Abstract
William White Howells was one of the intellectual giants of the discipline of biological anthropology during the twentieth century. He was a devoted student of Earnest A. Hooton; yet he played a central role in directing the discipline away from the typological thinking that infused the work of his predecessor, and toward the population perspective that characterizes the field today. An original and productive scholar with diverse interests, his influence was extraordinary not merely because of his brilliance, but also because of the kind of mentor he was. Almost two dozen graduate students, and countless others with whom he interacted in various capacities, have carried Howells' legacy into the twenty-first century.
Link
Differences between Early and Late Upper Paleolithic Europeans
Volume 137 Issue S47, Pages 70 - 99
Hunters of the Ice Age: The biology of Upper Paleolithic people
Brigitte M. Holt, Vincenzo Formicola
Abstract
The Upper Paleolithic represents both the phase during which anatomically modern humans appeared and the climax of hunter-gatherer cultures. Demographic expansion into new areas that took place during this period and the diffusion of burial practices resulted in an unprecedented number of well-preserved human remains. This skeletal record, dovetailed with archeological, environmental, and chronological contexts, allows testing of hypotheses regarding biological processes at the population level. In this article, we review key studies about the biology of Upper Paleolithic populations based primarily on European samples, but integrating information from other areas of the Old World whenever possible. Data about cranial morphology, skeletal robusticity, stature, body proportions, health status, diet, physical activity, and genetics are evaluated in Late Pleistocene climatic and cultural contexts. Various lines of evidence delineate the Last Glacial Maximum (LGM) as a critical phase in the biological and cultural evolution of Upper Paleolithic populations. The LGM, a long phase of climatic deterioration culminating around 20,000 BP, had a profound impact on the environment, lifestyle, and behavior of human groups. Some of these effects are recorded in aspects of skeletal biology of these populations. Groups living before and after the LGM, Early Upper Paleolithic (EUP) and Late Upper Paleolithic (LUP), respectively, differ significantly in craniofacial dimensions, stature, robusticity, and body proportions. While paleopathological and stable isotope data suggest good health status throughout the Upper Paleolithic, some stress indicators point to a slight decline in quality of life in LUP populations. The intriguing and unexpected incidence of individuals affected by congenital disorders probably indicates selective burial practices for these abnormal individuals. While some of the changes observed can be explained through models of biocultural or environmental adaptation (e.g., decreased lower limb robusticity following decreased mobility; changes in body proportions along with climatic change), others are more difficult to explain. For instance, craniodental and upper limb robusticity show complex evolutionary patterns that do not always correspond to expectations. In addition, the marked decline in stature and the mosaic nature of change in body proportions still await clarifications. These issues, as well as systematic analysis of specific pathologies and possible relationships between genetic lineages, population movements and cultural complexes, should be among the goals of future research.
Link
Later Stone Age South African Hunter gatherers and Herders as a single population
Journal of Archeological Science doi: 10.1016/j.jas.2008.11.001
Craniometric evidence for South African Later Stone Age herders and hunter-gatherers being a single biological population
Deano D. Stynder
Abstract
Later Stone Age (LSA) hunter-gatherers and herders co-existed in South Africa during the last 2,000 years. In spite of being the focus of intensive research over the years, the biological status and origins of the herders are still unclear. Did they represent a genetically distinct immigrant population who remained separate from the indigenous hunter-gatherers, or where they indigenous hunter-gatherers who took up herding after contact with herders, probably in northern Botswana? Here, this issue is investigated using craniometric data collected on a large sample of individually dated human crania from coastal LSA context. Mahalanobis distances (D), calculated from the raw metric data, show that there was a small increase in inter-individual craniofacial variation after the introduction of herding at c. 2,000 BP. Here it is argued that this small increase in variation is neither consistent with a largescale immigration of genetically distinct herders into South Africa, or the long-term coexistence of two genetically distinct populations. Two alternative explanations fit the data better: (1) herding entered South Africa via the small-scale immigration of genetically distinct herders; and (2) local hunter-gatherer populations adopted herding after coming in contact with herders in northern Botswana. While small-scale immigration would not have had a major influence on the local gene pool, it would have increased variation to some extent as immigrants mixed with local populations. If small-scale external gene flow was not a factor in the introduction of herding, secular issues related to the introduction of herding could explain the increased variation in post-2,000 BP populations.
Link
November 13, 2008
Warfare and human evolution
Now a new theory is emerging that challenges the prevailing view that warfare is a product of human culture and thus a relatively recent phenomenon. For the first time, anthropologists, archaeologists, primatologists, psychologists and political scientists are approaching a consensus. Not only is war as ancient as humankind, they say, but it has played an integral role in our evolution.Here is the website of this conference: Evolutionary Perspectives on War Conference. More from the New Scientist article:
...
These ideas emerged at a conference last month on the evolutionary origins of war at the University of Oregon in Eugene. "The picture that was painted was quite consistent," says Mark Van Vugt, an evolutionary psychologist at the University of Kent, UK. "Warfare has been with us for at least several tens, if not hundreds, of thousands of years."
Studies suggest that warfare accounts for 10 per cent or more of all male deaths in present-day hunter-gatherers. ... Primatologists have known for some time that organised, lethal violence is common between groups of chimpanzees, our closest relatives. ... Such raids are possible because humans and chimps, unlike most social mammals, often wander away from the main group to forage singly or in smaller groups, says Wrangham. ... Several participants presented the strongest evidence yet that males - whose larger and more muscular bodies make them better suited for fighting - have evolved a tendency towards aggression outside the group but cooperation within it. ... Aggression in women, she notes, tends to take the form of verbal rather than physical violence, and is mostly one on one.
...
Some of this behaviour could arguably be attributed to conscious mental strategies, but anthropologist Mark Flinn of the University of Missouri at Columbia has found that group-oriented responses occur on the hormonal level, too. He found that cricket players on the Caribbean island of Dominica experience a testosterone surge after winning against another village. But this hormonal surge, and presumably the dominant behaviour it prompts, was absent when the men beat a team from their own village, Flinn told the conference. "You're sort of sending the signal that it's play. You're not asserting dominance over them," he says. Similarly, the testosterone surge a man often has in the presence of a potential mate is muted if the woman is in a relationship with his friend. Again, the effect is to reduce competition within the group, says Flinn. "We really are different from chimpanzees in our relative amount of respect for other males' mating relationships."
...
Though women seem to help broker harmony within groups, says Van Vugt, men may be better at peacekeeping between groups.
November 12, 2008
Ancient DNA damage and phylogenetic histories
A way to deal with damage is to limit oneself to the study of genetic changes that are resilient to it, e.g., transversions over the commoner transitions. But, by doing so, the statistical power is diminished (less data is used). What the authors of this paper do, is try to use the full ancient DNA data, while accounting for the effects of damage in a systematic way. Their conclusions -for their test case- is that its overall effect is not very pronounced.
Molecular Biology and Evolution, doi:10.1093/molbev/msn256
Accommodating the effect of ancient DNA damage on inferences of demographic histories
Andrew Rambaut et al.
Abstract
DNA sequences extracted from ancient remains are increasingly used to generate large population data sets, often spanning tens of thousands of years of population history. Bayesian coalescent methods such as those implemented in the software package BEAST can be used to estimate the demographic history of these populations, sometimes resulting in complex scenarios of fluctuations in population size, which can be correlated with the timing of environmental events, such as glaciations. Recently, however, Axelsson et al. (2008) claimed that many of these complex demographic trends are likely to be the result of post-mortem DNA damage, a problem that they investigate by removing all sites involving transitions from ancient sequences prior to analysis. When this solution is applied to a previously published data set of Pleistocene bison, they show that the demographic signal of population expansion and decline disappears. While some apparently segregating mutations in ancient sequences may be due to post-mortem damage, we argue that discarding the data will result in loss of power to detect patterns of population change. Instead, to accommodate this problem, we implement a model in which sequences are the result of a joint process of molecular evolution and post-mortem DNA damage within a probabilistic inference framework. Through simulation, we demonstrate the ability of this model to accurately recover evolutionary parameters, demographic history and DNA damage rates. When this model is applied to the bison data set, we find that the rate of DNA damage is significant but low, and that the reconstruction of population size history is nearly identical to previously published estimates.
Link
November 11, 2008
Belgian Spy Neanderthals 36,000 years BP old
New data on the late Neandertals: Direct dating of the Belgian Spy fossils
Patrick Semal et al.
Abstract
In Eurasia, the period between 40,000 and 30,000 BP saw the replacement of Neandertals by anatomically modern humans (AMH) during and after the Middle to Upper Paleolithic transition. The human fossil record for this period is very poorly defined with no overlap between Neandertals and AMH on the basis of direct dates. Four new 14C dates were obtained on the two adult Neandertals from Spy (Belgium). The results show that Neandertals survived to at least ≈ 36,000 BP in Belgium and that the Spy fossils may be associated to the Lincombian-Ranisian-Jerzmanowician, a transitional techno-complex defined in northwest Europe and recognized in the Spy collections. The new data suggest that hypotheses other than Neandertal acculturation by AMH may be considered in this part of Europe.
Link
November 10, 2008
HGDP selection browser @ the Pritchard Lab
November 09, 2008
"War of the Gods" vs. "Clash of the Titans"
Variety reports:
Relativity Media is negotiating with Henry Cavill ("The Tudors") to star as Theseus in the Tarsem Singh-directed "War of the Gods."
At the same time, Warner Bros. is in talks with Sam Worthington to play the role of Perseus in the Louis Leterrier-directed "Clash of the Titans."
Deals for both actors are expected to be worked out, keeping the Greek mythology-themed projects on a parallel track. Both films are expected to begin production by late winter or early spring.
The projects have different plots, but each film will be made for under $100 million because the visual effects will be accomplished using the greenscreen techniques that made "300" so visually arresting.
Relativity got in the Greek game last summer when it bought the Charley and Vlas Parlapanides-scripted "War of the Gods" (Daily Variety, June 26).
Hollywood Gang's Gianni Nunnari and Canton Prods.' Mark Canton are producing with Ryan Kavanaugh.
At the same time, "The Incredible Hulk" helmer Leterrier committed to WB's "Clash of the Titans," a remake of the 1981 film that tells the story of Zeus son Perseus' journey and battles against Medusa. Scripted by Lawrence Kasdan, the WB film is a co-production with Legendary Pictures, produced by Basil Iwanyk of Thunder Road and Kevin De La Noy.
Cavill is best known for his work on Showtime's "The Tudors" but was also on the shortlist to play the Man of Steel in "Superman Returns."
Worthington, one of the finalists for the James Bond role that went to Daniel Craig, is starring for director James Cameron in "Avatar" and plays a pivotal role in the McG-directed "Terminator Salvation."
November 07, 2008
Sexual orientation of parents does affect their children
Psychol Rep. 2008 Aug;103(1):275-304.
Re-evaluation of the "no differences" hypothesis concerning gay and lesbian parenting as assessed in eight early (1979-1986) and four later (1997-1998) dissertations.
Schumm WR.
Academic and policy effects of eight early dissertations on gay and lesbian parenting are discussed with a focus on their having been cited at least 234 times in over 50 literature reviews, beginning with Gottman in 1989 and 1990. Most literature reviews, referencing these eight early dissertations and agreeing with Gottman's early conclusions, have reiterated the theme that parenting by gay men or lesbians has outcomes no different than parenting by heterosexual parents. Here it is proposed that certain potential adverse findings may have been obscured by suppressor effects which could have been evaluated had multivariate analyses been implemented. Further, several adverse findings were detected by reanalyzing data where sufficient information was yet available. Some of the dissertations' results (absent controls for social desirability and other differences between homosexual and heterosexual parents) supported the 2001 "no differences" hypothesis discussed by Stacey and Biblarz. Yet, differences were also observed, including some evidence in more recent dissertations, suggesting that parental sexual orientation might be associated with children's later sexual orientation and adult attachment style, among other outcomes. Odds ratios associated with some of the apparent effects were substantial in magnitude as well as statistically significant. Also, more recent research on gay and lesbian parenting continues to be flawed by many of the same limitations as previous research in this area of study, including overlooked suppressor effects.
Link
November 06, 2008
In search of the Hidden Heritability
So, if these traits are in our genes, how come we can't find them there?
The article does a great job at identifying the possible ways to find the "hidden heritability". Here they are, in my own words:
1. Look at more DNA spots
There is a long way between the million or so DNA bases covered by current microarray chips and the whole human genome. Because of linkage disequilibrium, i.e., DNA's propensity to be cut and inherited in large chunks, and not small pieces, you can often tell the value of a marker by looking at nearby markers. But, still, you don't really know until you look. So, denser microarrays, or even whole genome sequences may uncover some of the hidden heritability.
2. Look at more people
Associations between traits and genes are established by statistics. To find a weak association, or an association between a not-so-common variant and the trait in question, you need a large sample. So, if the hidden heredity is hidden away in markers that are beyond your statistical power, you can simply increase this power: sample more people.
3. Look at copy-number variations (CNVs)
Any two individuals don't just have single-letter differences, but also structural changes, where an individual may have more or fewer copies of entire chunks of DNA. So, by looking at single nucleotides you are examining one source of human variation, but missing another chunk of it that may as important.
3. Study gene-gene interactions
Genes form complex networks of interaction. If you flip a SNP from C to T, you don't always get the same effect on the phenotype. This flip may increase, decrease, or leave unaffected, your risk for a disease, depending on what other genes you have. This epistatic interaction of genes makes it difficult to detect associations. It's a lot easier to study the individual effects of 2N alleles at N genes than it is to study the effects of 2N possible combinations.
4. Don't trust heritability estimates
What if inherited conditions thought to be genetic aren't really genetic, because of epigenetic modifications of gene expression, or shared environments (e.g., in the womb) that aren't accounted for?
5. Don't trust diagnoses of conditions
If you want to find a correlation between a gene G and a trait T, you'd better be sure what T actually is. If it's a whole set of different behaviors, conveniently bundled into a condition T (such as schizophrenia), then you're in trouble, since each of these conditions may have its own causative agent. Many major diseases may be caused by more than one underlying condition, with a different genetic background. So, if you are seeking to find the common thread between people with trait T, you might not find it because there is no common thread!
My guess is that the bulk of the missing heritability is to be found in three sources:
- Epistasis. Humans are makeshift accidents of evolution, and not well-engineered machines where the effects of individual components have been designed to work well in isolation, shielding other components from their effects. Most things in the human body affects most other things, either directly or indirectly. There are, of course, some master switches which do have individual pronounced effects (e.g., giving you lactose tolerance or breast cancer), but these are the exception. Normal variation is due to how well-put together the individual is, and not so much in the individual components.
- Gene-Environment interactions. Just as the effect of genes depends on the joint presence of other genes (epistasis), so it depends on the presence of particular environmental influences. Imagine an allele that shows zero association with a particular trait. Does this mean that it has no influence on that trait? No, since zero association is perfectly compatible with even a huge influence, provided that a positive influence under one type of genomic or environmental background is balanced by a negative influence under another.
- Very low frequency (family) alleles. Natural selection faces a constant battle against the continuing re-emergence of less-than-optimal alleles. Children are almost certainly on average genetically worse than their parents, since parents have survived and reproduced, while children's ability to do so is yet to be tested (*) While human variation is -in part- due to long-lived alleles that have braved the generations, quite a lot of it is due to recent alleles that arose in families, and have not had the time to spread to many bodies. It is these extremely rare family alleles and allele combinations that population studies can't quite capture.
Some related posts on the limits of genome-wide association studies: on intelligence, on height and body mass index, and on CNVs.
(*) Incidentally, this is why the population replacement rate is more than 2 children per woman.
November 05, 2008
Time-dependency of the human mtDNA evolutionary mutation rate
I had planned to write a post titled: "On the difficulty of archaeological calibration of the mutation rate", for some time. My goal was to follow on my criticism of the proposed explanation for a supposedly lower evolutionary rate, by a criticism of the alleged fact that such a lower rate is proven by archaeological calibration.
So, I was pleasantly surprised to see a new paper which allows me to frame my thoughts in a concrete context. The new paper's purpose is precisely this: to calibrate the evolutionary mutation rate archaeologically.
PREAMBLE
A molecular clock typically works according to this generic equation:
VARIATION = TIME * MUTATION RATE
Some measure of VARIATION is obtained in the present time (e.g. the ρ or π statistics in this paper), and some estimate of the MUTATION RATE is established. This allows us to calculate an estimate of TIME.
How is the MUTATION RATE established? Either by direct measurement, i.e., by looking at relatives and seeing how their genotypes actually differ from each other, or by calibration whereby present-day VARIATION is measured for events with (supposedly) known TIME, leading to an estimate of the MUTATION RATE as VARIATION/TIME.
"KNOWN" TIME?
Two typical examples of calibration go like this:
- Humans and Chimps split T millions of years ago. The amount of human-chimp differentiation is D. Therefore, the human-chimp divergence rate was on average D/T over the time period of T years.
- The ancestors of Native Americans arrived T thousands of years ago. Variation within one of their founding lineages (e.g., mtDNA haplogroup D1) is D. Therefore, variation increased by a rate of D/T over the time period of T years.
But, consider more well-established archaeological events like the arrival of the Native Americans which is based on carbon dating and surveys of many sites across the continent. This, apparently gives us a secure T. Or does it?
There are at least two reasons why it does not: demography and selection.
The arrival of Europeans in the new World is perfectly known: it started in 1492. Pre-existing migrants such as the Vikings do not appear to have made a lasting contribution. Yet, if we calculate diversity within European-origin haplogroups in the New World, we will find that they are pretty much as diverse as they are in Europe, Why? Because the migration involved a large number of migrants.
If, a few thousand years from now, after a collapse and rebirth of human civilization, geneticists look at the genes of Americans of that future time, they might conclude that Amerindians and Europeans arrived to the continent at the same time, or even that lineages of the latter (e.g., mtDNA haplogroup U5) preceded those of the former (e.g., mtDNA haplogroup D1).
But, suppose that a limited number of migrants arrive at the archaeologically calibrated time, i.e., a "founder effect", e.g., a single mtDNA D1 "mother" arrived in the New World T years ago.
How do we know that she is the most-recent-common anestor of present-day D1 women from the New World?
We know that she is a common ancestor, but not necessarily the most recent common ancestor (MRCA). Indeed, if selection is at play, then particular lineages overwhelm those of their relatives, and an D1 woman with an advantageous lineage, who lived long after the first D1 woman, may be the real MRCA.
Thus, archaeological calibration is often an illusion: lineages may appear to be older or younger than the calibration time, depending on the population's demography and the effects of selection.
THE NON-EXISTING CONSENSUS
The authors of this study take for granted that there is a discrepancy between the mutation rate created by measurement and calibration (germline vs. evolutionary). Yet, a recent paper paints quite a different picture, finding no difference between the two rates. How do they arrive at such a conclusion? By looking directly at the gene pool of a population in two different points in time (using ancient DNA), and not relying on calibration.
TIME-DEPENDENCY OF THE MUTATION RATE
These criticisms aside, the current paper does have some important implications about the evolutionary rate. Its central idea is that the molecular clock works like this:
VARIATION = TIME * MUTATION RATE(TIME)
i.e., that the MUTATION RATE is itself a function of time. The above equation, like its previous simpler version allows us to estimate TIME from VARIATION. But, it introduces an additional complication. How does the evolutionary mutation rate vary across different time scales?
This, in itself, is a step in the right direction. For example, in human Y-chromosomes a slower evolutionary rate was proposed by Zhivotovsky et al. (2004) (pdf) based on ~1,000 year histories of Bulgarian Gypsies and Polynesians, and a very different rate was proposed by Forster et al. (2000), using a different calibration based on Native American prehistory. Yet, the more widely used Zhivotovsky rate has been used to age 2,700 or 60,000-year-old haplogroups, as if it was equally applicable in both time scales.
The authors of this paper make some interesting comments:
Genealogy-based rate estimates between 2,500-5,000ya are indistinguishable from pedigree-based mutation rate estimate (tables 1-2).So, for events of the recent past, to at least the Bronze Age, "pedigree" and "evolutionary" rates are indistinguishable. Lineages that appear to be of Bronze Age origin based on the pedigree rate, are indeed that old, and not Neolithic or Paleolithic as might be predicted by use of a slower "evolutionary" rate.
Continuing:
The time period between 5,000ya and ~15,000ya represents both a break in our dataset and a sudden decline in estimated mutation rate (fig. 2). Estimates for the intermediate period roughly 15,000ya are about 40% lower than estimates calibrated on arrival dates less than 5,000yaThis is a break in their dataset because, well, they don't have any calibration points between 5-15ky. Thus, their estimate is nothing more than an extrapolation based on calibration points younger than 5ky or older than 15ky. But still, their estimates for even 15ky (which upper bounds all the Neolithic and Mesolithic for humans) are only 40% lower than using pedigree rates. Thus, even if we accept this extrapolation, ages between 5 and 15ky using the pedigree rate may be somewhat underestimated, but certainly not by a huge factor.
Continuing:
Using simulation, Zhivotovsky, Underhill and Feldman (2006) showed that microsatellite mutation rates estimated from small populations (haplogroups) undergoing serial bottlenecks are indeed reduced compared to pedigree-based rates.
The key word here is small, which is why using such reduced rates for haplogroups that consist of tens of millions of men is nonsense.
In simple English, they propose that the evolutionary mutation rate for large Neolithic populations was close to the pedigree rate, while for the smaller pre-Neolithic hunter-gatherer groups it was much lower.
The striking difference between our mutation rate estimates from before and after 20,000 years ago suggests that demographic history may play an important role. Prior to the Last Glacial Maximum lasting between 22,000-15,000ya, human populations were characterized by small, mobile hunter-gatherer groups that may have been frequently subject to fluctuations in population size. Following the LGM, humans experienced far fewer climatic swings (Mithen 2004). Particularly after the Younger Dryas cycle, agriculture facilitated dramatic population growth and serial bottlenecks were unlikely to substantially reduce the diversity of such large populations. A dramatic increase in population size during the Neolithic period is supported by mtDNA genomes from African, southeastern Asian, and European populations (Gignoux C, Henn B, unpublished data). We propose that a population history consisting of serial bottlenecks followed by recent population growth currently provides the most compelling explanation for the time-dependency of human hypervariable and coding region mtDNA mutation rate estimates.
Which is pretty much what I've been saying for the last few months:
But, if you read Zhivotovsky, Underhill and Feldman (2006) or my two previous posts on the subject, you will realize that the effective rate depends on population history; that the 0.00069 rate is derived for constant-sized populations where haplogroups never grow to large numbers.or:
Of course, if one studies numerically small populations, it is possible that a slower effective rate may be desired. My concern is with the large human populations (e.g. Greeks or Indians) where real haplogroup sizes exceed greatly those produced by simulations with reproductive equality.or:
Z.U.F. have also proposed two additional demographic scenaria under which a higher effective mutation rate would be observed:Let's hope that -if nothing else- this paper will be the beginning of the end for the indiscriminate use of "evolutionary rates" across different time spans and marker systems. I am not counting on this happening any time soon, given the substantial intellectual inertia of the field.Both factors seem reasonable for post-Holocene human populations. It is well known that -whatever temporary setbacks there were- mankind has overall experienced a substantial population growth in recent millennia. Thus, an expanding population seems like a fair assumption.
- A sudden jump in the size of the haplogroup after it appears
- An expanding population (m>1)
CONCLUSION
I have expressed my reservations about the difficulty of "archaeological calibration" of the mutation rate. These thoughts pertain to the difficulty of obtaining a valid calibration point due to demography and selection. They also find indirect support from calibration of the mutation rate via ancient DNA.
Nonetheless, even within a calibrationist framework, this paper shows that human mtDNA over the last 5,000 years has accumulated variation at the germline (pedigree) mutation rate, and has extrapolated that in the last 15ky at something close to it -- and definitely not at a much slower rate.
Hopefully, this paper will be extended to rethink the calibration of human Y-chromosome and autosomal mutation rates, and its assumptions may be checked from ancient DNA-based calibration in both humans and other species.
Molecular Biology and Evolution doi: 10.1093/molbev/msn244
Characterizing the Time-Dependency of Human Mitochondrial DNA Mutation Rate Estimates
Brenna M. Henn et al.
Abstract
Previous research has established a discrepancy of nearly an order of magnitude between pedigree-based and phylogeny-based (human vs. chimpanzee) estimates of the mitochondrial (mtDNA) control region mutation rate. We characterize the time-dependency of the human mitochondrial hypervariable region one (HVRI) mutation rate by generating fourteen new phylogeny-based mutation rate estimates using within-human comparisons and archaeological dates. Rate estimates based on population events between 15,000 and 50,000 years ago are at least twofold lower than pedigree-based estimates. These within-human estimates are also higher than estimates generated from phylogeny-based human-chimpanzee comparisons. Our new estimates establish a rapid decay in evolutionary mutation rate between approximately 2,500 and 50,000 years ago, and a slow decay from 50,000 to 6 million years ago. We then extend this analysis to the mtDNA coding region. Our within-human coding region mutation rate estimates display a similar, though less rapid, time-dependent decay. We explore the possibility that multiple hits explain the discrepancy between pedigree-based and phylogeny-based mutation rates. We conclude that while nucleotide substitution models incorporating multiple-hits do provide a possible explanation for the discrepancy between pedigree-based and human-chimpanzee mutation rate estimates, they do not explain the rapid decline of within-human rate estimates. We propose that demographic processes such as serial bottlenecks prior to the Holocene could explain the difference between rates estimated before and after 15,000 years ago. Our findings suggest that human mitochondrial DNA estimates of dates of population and phylogenetic events should be adjusted in light of this time-dependency of the mutation rate estimates.
Link
