A new paper has appeared about Y chromosome haplogroups in Crete. I will read it more carefully and comment later on. I would be interested in hearing if there are any traditions about the people from Lasithi, which could be correlated with the genetic evidence. Also, I do not have Poulianos' "Origins of the Cretans" books handy, which covers quite extensively the physical anthropology of Crete, so feel free to leave a comment with any relevant information.
A first comment based on a reading of the abstract would be that the similarity between R1b chromosomes from Lasithi and NE Italy could be a legacy of the Venetian period of Cretan history.
UPDATE 1: Absence of non-Caucasoid influences in Lasithi and Heraklion.
There is an absence of Sub-Saharan Y chromosomes (A, B, or E*(xE3b)) in Lasithi and Herakleio. Mongoloid Y chromosomes are also absent. These would be represented by haplogroups C, N, and O. C was typed directly, and the absence of N and O can be inferred from the absence of K* chromosomes using the markers tested. The only chromosomes that could be of non-Caucasoid origin are 2 Q chromosomes, which are more typical of Siberian and Native American populations, although they have been detected in trace amounts in the Middle East and Europe. It remains to be seen whether or not these Q chromosomes represent prehistoric remnants or more recent arrivals from Asia.
UPDATE 2: Limited occurrence of J1 signifies absence of significant Arab influences in Lasithi and Heraklion.
Arab and other Semitic populations usually possess an excess of J1 Y chromosomes compared to other populations harboring Y-haplogroup J. As I have noted elsewhere the predominance of J2 over J1 is typical of European populations of J. In the Cretan samples there are 65 J2 chromosomes and 3 J1 chromosomes. It should be noted that these may not necessarily be of Arab or Semitic origin, since J1 is also found at some frequency in the northern parts of the Middle East inhabited by Caucasian and Iranic speakers and may have been present in early Neolithic times. In any case, the preponderance of J2 over J1 and low occurrence of J1 suggests that the Arab rule of Crete had little effect in terms of characteristically Arab Y chromosomes.
European Journal of Human Genetics advance online publication 31 January 2007
Paleolithic Y-haplogroup heritage predominates in a Cretan highland plateau
Laisel Martinez1, Peter A Underhill2, Lev A Zhivotovsky3, Tenzin Gayden1, Nicholas K Moschonas4, Cheryl-Emiliane T Chow2, Simon Conti2, Elisabetta Mamolini5, L Luca Cavalli-Sforza2 and Rene J Herrera1
Abstract
The island of Crete, credited by some historical scholars as a central crucible of western civilization, has been under continuous archeological investigation since the second half of the nineteenth century. In the present work, the geographic stratification of the contemporary Cretan Y-chromosome gene pool was assessed by high-resolution haplotyping to investigate the potential imprints of past colonization episodes and the population substructure. In addition to analyzing the possible geographic origins of Y-chromosome lineages in relatively accessible areas of the island, this study includes samples from the isolated interior of the Lasithi Plateau – a mountain plain located in eastern Crete. The potential significance of the results from the latter region is underscored by the possibility that this region was used as a Minoan refugium. Comparisons of Y-haplogroup frequencies among three Cretan populations as well as with published data from additional Mediterranean locations revealed significant differences in the frequency distributions of Y-chromosome haplogroups within the island. The most outstanding differences were observed in haplogroups J2 and R1, with the predominance of haplogroup R lineages in the Lasithi Plateau and of haplogroup J lineages in the more accessible regions of the island. Y-STR-based analyses demonstrated the close affinity that R1a1 chromosomes from the Lasithi Plateau shared with those from the Balkans, but not with those from lowland eastern Crete. In contrast, Cretan R1b microsatellite-defined haplotypes displayed more resemblance to those from Northeast Italy than to those from Turkey and the Balkans.
Link
January 31, 2007
Tree of life may not capture the complexities of evolution
The tree of life concept basically states that all living organisms are descended from previous living organisms, all the way to a unique common ancestor. However, this concept does not capture the possibility of lateral gene transfer, i.e., the fact that genes aren't simply inherited from one's ancestor(s) within the same species but can be transferred across species. The authors make the good point that algorithms designed to infer tree structures from genetic data, will -no surprise- infer such structures, and hence do not really "support" the idea that such structures really fit the evolutionary picture.
Proc. Natl. Acad. Sci. USA, 10.1073/pnas.0610699104
Pattern pluralism and the Tree of Life hypothesis
W. Ford Doolittle and Eric Bapteste
Darwin claimed that a unique inclusively hierarchical pattern of relationships between all organisms based on their similarities and differences [the Tree of Life (TOL)] was a fact of nature, for which evolution, and in particular a branching process of descent with modification, was the explanation. However, there is no independent evidence that the natural order is an inclusive hierarchy, and incorporation of prokaryotes into the TOL is especially problematic. The only data sets from which we might construct a universal hierarchy including prokaryotes, the sequences of genes, often disagree and can seldom be proven to agree. Hierarchical structure can always be imposed on or extracted from such data sets by algorithms designed to do so, but at its base the universal TOL rests on an unproven assumption about pattern that, given what we know about process, is unlikely to be broadly true. This is not to say that similarities and differences between organisms are not to be accounted for by evolutionary mechanisms, but descent with modification is only one of these mechanisms, and a single tree-like pattern is not the necessary (or expected) result of their collective operation. Pattern pluralism (the recognition that different evolutionary models and representations of relationships will be appropriate, and true, for different taxa or at different scales or for different purposes) is an attractive alternative to the quixotic pursuit of a single true TOL.
Link
Proc. Natl. Acad. Sci. USA, 10.1073/pnas.0610699104
Pattern pluralism and the Tree of Life hypothesis
W. Ford Doolittle and Eric Bapteste
Darwin claimed that a unique inclusively hierarchical pattern of relationships between all organisms based on their similarities and differences [the Tree of Life (TOL)] was a fact of nature, for which evolution, and in particular a branching process of descent with modification, was the explanation. However, there is no independent evidence that the natural order is an inclusive hierarchy, and incorporation of prokaryotes into the TOL is especially problematic. The only data sets from which we might construct a universal hierarchy including prokaryotes, the sequences of genes, often disagree and can seldom be proven to agree. Hierarchical structure can always be imposed on or extracted from such data sets by algorithms designed to do so, but at its base the universal TOL rests on an unproven assumption about pattern that, given what we know about process, is unlikely to be broadly true. This is not to say that similarities and differences between organisms are not to be accounted for by evolutionary mechanisms, but descent with modification is only one of these mechanisms, and a single tree-like pattern is not the necessary (or expected) result of their collective operation. Pattern pluralism (the recognition that different evolutionary models and representations of relationships will be appropriate, and true, for different taxa or at different scales or for different purposes) is an attractive alternative to the quixotic pursuit of a single true TOL.
Link
"Hobbit" wars continue
From the Toronto Star:
In the ongoing battle between the two camps, the latest evidence is in.
The hobbit is definitely a new species of human, related to but separate from Homo sapiens, concludes a study by a Florida State University team published yesterday in the Proceedings of the National Academy of Sciences. Neuro-paleontologist Dean Falk, the head researcher, says she is "absolutely convinced" the brain of LB1, as the hobbit is officially known, is not abnormal.
"It isn't microcephalic in any way," she told the Star. "Furthermore, its features are the antithesis of that condition."
Because the brain leaves an image imprinted on the skull, its shape can be reconstructed into a three-dimensional, computer-generated model called an endocast. Falk's team compared the endocasts of 21 skulls; 10 of normal humans, nine of people with microcephaly, one a dwarf and LB1. Using statistical techniques, they classified them into normal and microcephalic.
The hobbit's brain – one-third the size of a modern human's – fell clearly within the normal range, says Falk. Furthermore, its brain has four unusual features, including large frontal and temporal lobes, which distinguishes it from Homo sapiens, she says, and justifies its classification as a separate species, Homo floresiensis.
Falk had expected to see something that looked like the brain of a chimpanzee, but says LB1's brain is sophisticated and unique, unlike other primate brain casts she has seen.
"It is a little, but highly evolved, brain that had been globally reorganized. It didn't get bigger, it got more complex. It got rewired and reorganized."
January 25, 2007
mtDNA Differences between populations of Ashkenazi Jews
From the paper:
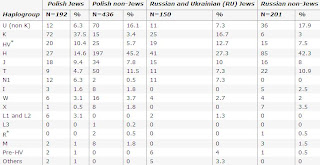
Eur J Hum Genet. 2007 Jan 24; [Epub ahead of print]
Ashkenazi Jewish mtDNA haplogroup distribution varies among distinct subpopulations: lessons of population substructure in a closed group.
Feder J, Ovadia O, Glaser B, Mishmar D.
The quest for genes associated with diseases is widely recognized as an essential task in the effort to investigate the genetic basis of complex human disorders and traits. A basic stage in association studies is the careful choice of the model population, with preference to closed groups having little population substructure. Here, we show evidence for significant geographic substructure (P=0.017) of the maternal lineage represented by mitochondrial DNA variation in one of the most commonly studied populations, the Ashkenazi Jews. Most of the substructure effect stems from differential representation of haplogroups K and H. Our results underline the essentiality of adjusting data of population genetic variation for substructure during the design of association studies, even in apparently closed populations.
Link

Eur J Hum Genet. 2007 Jan 24; [Epub ahead of print]
Ashkenazi Jewish mtDNA haplogroup distribution varies among distinct subpopulations: lessons of population substructure in a closed group.
Feder J, Ovadia O, Glaser B, Mishmar D.
The quest for genes associated with diseases is widely recognized as an essential task in the effort to investigate the genetic basis of complex human disorders and traits. A basic stage in association studies is the careful choice of the model population, with preference to closed groups having little population substructure. Here, we show evidence for significant geographic substructure (P=0.017) of the maternal lineage represented by mitochondrial DNA variation in one of the most commonly studied populations, the Ashkenazi Jews. Most of the substructure effect stems from differential representation of haplogroups K and H. Our results underline the essentiality of adjusting data of population genetic variation for substructure during the design of association studies, even in apparently closed populations.
Link
January 24, 2007
Sub-Saharan African Y chromosome haplogroup A in white British surname
From the BBC story:
[UPDATE] From the paper:
Here is the abstract:
European Journal of Human Genetics (advance online publication)
Africans in Yorkshire? The deepest-rooting clade of the Y phylogeny within an English genealogy
Turi E King et al.
Abstract
The presence of Africans in Britain has been recorded since Roman times, but has left no apparent genetic trace among modern inhabitants. Y chromosomes belonging to the deepest-rooting clade of the Y phylogeny, haplogroup (hg) A, are regarded as African-specific, and no examples have been reported from Britain or elsewhere in Western Europe. We describe the presence of an hgA1 chromosome in an indigenous British male; comparison with African examples suggests a Western African origin. Seven out of 18 men carrying the same rare east-Yorkshire surname as the original male also carry hgA1 chromosomes, and documentary research resolves them into two genealogies with most-recent-common-ancestors living in Yorkshire in the late 18th century. Analysis using 77 Y-short tandem repeats (STRs) is consistent with coalescence a few generations earlier. Our findings represent the first genetic evidence of Africans among 'indigenous' British, and emphasize the complexity of human migration history as well as the pitfalls of assigning geographical origin from Y-chromosomal haplotypes.
Link
People of African origin have lived in Britain for centuries, according to genetic evidence.From the EurekAlert release:
A Leicester University study found that seven men with a rare Yorkshire surname carry a genetic signature previously found only in people of African origin.
The men seem to have shared a common ancestor in the 18th Century, but the African DNA lineage they carry may have reached Britain centuries earlier.
...
By chance, the researchers discovered a white man with a rare Yorkshire surname carrying a Y chromosome haplogroup that had previously been found only in West African men. And even there, it is relatively uncommon.
"We found that he was in haplogroup A1, which is highly West African-specific," said Turi King, a co-author on the study at the University of Leicester.
"It is incredibly rare, there are only 25 other people known worldwide and they are all African."
...
Prior to the 20th Century, there have been various routes by which people of African ancestry might have reached Britain. For example, the Romans recruited from Africa and elsewhere for the garrison that guarded Hadrian's Wall.
...
Another major route was through the slave trade.
"Some of the Africans who arrived in Britain through the slave trade rose quite high up in society, and we know they married with the rest of the population," said Ms King.
"It could be either of these two routes," she said. Even if the two family trees link up in the 18th Century, haplogroup A1 could have reached Britain long before that.
"But my guess is that, because many slaves came from West Africa, it could have been through that route," Ms King told BBC News.
New research has identified the first genetic evidence of Africans having lived amongst "indigenous" British people for centuries. Their descendants, living across the UK today, were unaware of their black ancestry.I don't see the EJHG article on the website yet, feel free to leave a comment if I missed it or it is published online.
...
"As you can imagine, we were pretty amazed to find this result in someone unaware of having any African roots," explains Professor Jobling, a Wellcome Trust Senior Research Fellow. "The Y chromosome is passed down from father to son, so this suggested that Mr. X must have had African ancestry somewhere down the line. Our study suggests that this must have happened some time ago."
Although most of Britain's one million people who define themselves as "Black or Black British" owe their origins to immigration from the Caribbean and Africa from the mid-twentieth century onwards, in reality, there has been a long history of contact with Africa. Africans were first recorded in the north 1800 years ago, as Roman soldiers defending Hadrian's Wall.
To investigate the origins of hgA1 in Britain, the team recruited and studied a further eighteen males with the same surname as Mr. X. All but one were from the UK, with paternal parents and grandparents also born in Britain. Six, including one male in the US whose ancestors had migrated from England in 1894, were found to have the hgA1 chromosome.
...
"This study shows that what it means to be British is complicated and always has been," says Professor Jobling. "Human migration history is clearly very complex, particularly for an island nation such as ours, and this study further debunks the idea that there are simple and distinct populations or 'races'." [DP: This is a common fallacious argument against the existence of biological races; demand that races be completely genetically distinct, and then "debunk" their existence at any slightest sign of intermixture. It is indeed the distinctness of the European and Sub-Saharan African Y chromosome pools that allows us to infer the origin of rare outliers. Human races are not isolated islands, but rather more or less sharply defined hills or mountains on the genetic landscape].
In addition, Professor Jobling believes that the research may have implications for DNA profiling in criminal investigations.
"Forensic scientists use DNA analysis to predict a person's ethnic origins, for example from hair or blood samples found at a crime scene. Whilst they are very likely to predict the correct ethnicity by using wider analysis of DNA other than the Y chromosome, finding this remarkable African chromosome would certainly have them scratching their heads for a while."
[UPDATE] From the paper:
Our study shows that a globally rare Y-chromosome type, belonging to the deepest-rooting African branch of the Y-phylogeny, has been present in Northern England since at least the mid-18th century. HgE3a is by far the most frequent Y-chromosomal lineage in Africa, existing at 48% in a continent-wide sample of 1122 chromosomes,30 so we would expect any substantial past immigration from Africa to Britain to have left examples of chromosomes belonging to this common hg. However, a survey of 1772 Y chromosomes from the British Isles found none,13 and they are also absent from our control sample of 421 chromosomes. The general rarity of African lineages may reflect a low level of initial introgression, later loss through drift, or sampling bias – for example, the large British survey13 sampled from small towns, in which the descendants of early British Africans, who were concentrated in cities, may be depleted.
Admixture between populations of African and European origin is often sex-biased, with a greater proportion of the African component of the hybrid population being contributed by females.48 Assuming an equal number of males and females of African origin migrating to Britain, we might therefore expect mitochondrial DNA to reveal a stronger signal of African admixture than the Y chromosome. There is little published evidence, but a study of mitochondrial DNA sequence diversity among 100 'white Caucasian' British49 does contain one haplotype, which represents an hg L1c sequence (defined according to Salas et al.50), with a probable origin in West Central Africa.51 This could represent a possible maternal counterpart to the Y-lineage we describe here.
Here is the abstract:
European Journal of Human Genetics (advance online publication)
Africans in Yorkshire? The deepest-rooting clade of the Y phylogeny within an English genealogy
Turi E King et al.
Abstract
The presence of Africans in Britain has been recorded since Roman times, but has left no apparent genetic trace among modern inhabitants. Y chromosomes belonging to the deepest-rooting clade of the Y phylogeny, haplogroup (hg) A, are regarded as African-specific, and no examples have been reported from Britain or elsewhere in Western Europe. We describe the presence of an hgA1 chromosome in an indigenous British male; comparison with African examples suggests a Western African origin. Seven out of 18 men carrying the same rare east-Yorkshire surname as the original male also carry hgA1 chromosomes, and documentary research resolves them into two genealogies with most-recent-common-ancestors living in Yorkshire in the late 18th century. Analysis using 77 Y-short tandem repeats (STRs) is consistent with coalescence a few generations earlier. Our findings represent the first genetic evidence of Africans among 'indigenous' British, and emphasize the complexity of human migration history as well as the pitfalls of assigning geographical origin from Y-chromosomal haplotypes.
Link
January 21, 2007
Old people like arched eyebrows
You learn something new every day...
Aesthetic Plast Surg. 2007 Jan 17; [Epub ahead of print]
Attractiveness of Eyebrow Position and Shape in Females Depends on the Age of the Beholder.
Feser DK, Grundl M, Eisenmann-Klein M, Prantl L.
BACKGROUND: Great diversity exists among individuals with respect to eyebrow position and shape, and the notion of an "ideal" eyebrow has changed quite significantly over the past several decades. METHODS: This study compared three different variations of eyebrows. One variation was the arched eyebrow with the maximum height in the middle. The other two variations had their maximum height in the lateral third, but differed in their position (high vs low). For each of the seven female portraits presented, three variations were generated using morphing software. A total of 357 subjects 12 to 85 years of age compared these variations and ranked each woman individually with respect to perceived attractiveness. RESULTS: The data show that the preference for a specific eyebrow shape depends on a person's age. Young subjects up to 30 years of age preferred eyebrows in a lower position, and ruled out arched eyebrows. Subjects older than 50 years stated exactly the opposite preference. CONCLUSION: First, there is not one single beauty ideal for eyebrows, but at least three. The ideal a person prefers depends on his or her age. Second, because trends are generally introduced by young people and not by older individuals, and the young tend to prefer eyebrows in a lower position, it seems plausible to assume that the trend currently appears to be moving away from arched eyebrows toward lower positioned eyebrows with a maximum height in the lateral third.
Link
Aesthetic Plast Surg. 2007 Jan 17; [Epub ahead of print]
Attractiveness of Eyebrow Position and Shape in Females Depends on the Age of the Beholder.
Feser DK, Grundl M, Eisenmann-Klein M, Prantl L.
BACKGROUND: Great diversity exists among individuals with respect to eyebrow position and shape, and the notion of an "ideal" eyebrow has changed quite significantly over the past several decades. METHODS: This study compared three different variations of eyebrows. One variation was the arched eyebrow with the maximum height in the middle. The other two variations had their maximum height in the lateral third, but differed in their position (high vs low). For each of the seven female portraits presented, three variations were generated using morphing software. A total of 357 subjects 12 to 85 years of age compared these variations and ranked each woman individually with respect to perceived attractiveness. RESULTS: The data show that the preference for a specific eyebrow shape depends on a person's age. Young subjects up to 30 years of age preferred eyebrows in a lower position, and ruled out arched eyebrows. Subjects older than 50 years stated exactly the opposite preference. CONCLUSION: First, there is not one single beauty ideal for eyebrows, but at least three. The ideal a person prefers depends on his or her age. Second, because trends are generally introduced by young people and not by older individuals, and the young tend to prefer eyebrows in a lower position, it seems plausible to assume that the trend currently appears to be moving away from arched eyebrows toward lower positioned eyebrows with a maximum height in the lateral third.
Link
January 18, 2007
Charles Murray's Intelligence Trilogy in WSJ
Intelligence in the Classroom (Jan. 16)
Half of all children are below average, and teachers can do only so much for them.
Too many Americans are going to college.
Aztecs vs. Greeks (Jan. 18)
Those with superior intelligence need to learn to be wise.
Half of all children are below average, and teachers can do only so much for them.
Hardly anyone will admit it, but education's role in causing or solving any problem cannot be evaluated without considering the underlying intellectual ability of the people being educated.What's Wrong With Vocational School? (Jan. 17)
Too many Americans are going to college.
Put another way, it makes sense for only about 15% of the population, 25% if one stretches it, to get a college education. And yet more than 45% of recent high school graduates enroll in four-year colleges.
Aztecs vs. Greeks (Jan. 18)
Those with superior intelligence need to learn to be wise.
All of the above are antithetical to the mindset that prevails in today's schools at every level. The gifted should not be taught to be nonjudgmental; they need to learn how to make accurate judgments. They should not be taught to be equally respectful of Aztecs and Greeks; they should focus on the best that has come before them, which will mean a light dose of Aztecs and a heavy one of Greeks. The primary purpose of their education should not be to let the little darlings express themselves, but to give them the tools and the intellectual discipline for expressing themselves as adults.
Positive selection for human skin color
Annals of Human Genetics (OnlineEarly Articles)
Signatures of Positive Selection in Genes Associated with Human Skin Pigmentation as Revealed from Analyses of Single Nucleotide Polymorphisms
O. Lao et al.
Summary
Phenotypic variation between human populations in skin pigmentation correlates with latitude at the continental level. A large number of hypotheses involving genetic adaptation have been proposed to explain human variation in skin colour, but only limited genetic evidence for positive selection has been presented. To shed light on the evolutionary genetic history of human variation in skin colour we inspected 118 genes associated with skin pigmentation in the Perlegen dataset, studying single nucleotide polymorphisms (SNPs), and analyzed 55 genes in detail. We identified eight genes that are associated with the melanin pathway (SLC45A2, OCA2, TYRP1, DCT, KITLG, EGFR, DRD2 and PPARD) and presented significant differences in genetic variation between Europeans, Africans and Asians. In six of these genes we detected, by means of the EHH test, variability patterns that are compatible with the hypothesis of local positive selection in Europeans (OCA2, TYRP1 and KITLG) and in Asians (OCA2, DCT, KITLG, EGFR and DRD2), whereas signals were scarce in Africans (DCT, EGFR and DRD2). Furthermore, a statistically significant correlation between genotypic variation in four pigmentation candidate genes and phenotypic variation of skin colour in 51 worldwide human populations was revealed. Overall, our data also suggest that light skin colour is the derived state and is of independent origin in Europeans and Asians, whereas dark skin color seems of unique origin, reflecting the ancestral state in humans.
Link
Signatures of Positive Selection in Genes Associated with Human Skin Pigmentation as Revealed from Analyses of Single Nucleotide Polymorphisms
O. Lao et al.
Summary
Phenotypic variation between human populations in skin pigmentation correlates with latitude at the continental level. A large number of hypotheses involving genetic adaptation have been proposed to explain human variation in skin colour, but only limited genetic evidence for positive selection has been presented. To shed light on the evolutionary genetic history of human variation in skin colour we inspected 118 genes associated with skin pigmentation in the Perlegen dataset, studying single nucleotide polymorphisms (SNPs), and analyzed 55 genes in detail. We identified eight genes that are associated with the melanin pathway (SLC45A2, OCA2, TYRP1, DCT, KITLG, EGFR, DRD2 and PPARD) and presented significant differences in genetic variation between Europeans, Africans and Asians. In six of these genes we detected, by means of the EHH test, variability patterns that are compatible with the hypothesis of local positive selection in Europeans (OCA2, TYRP1 and KITLG) and in Asians (OCA2, DCT, KITLG, EGFR and DRD2), whereas signals were scarce in Africans (DCT, EGFR and DRD2). Furthermore, a statistically significant correlation between genotypic variation in four pigmentation candidate genes and phenotypic variation of skin colour in 51 worldwide human populations was revealed. Overall, our data also suggest that light skin colour is the derived state and is of independent origin in Europeans and Asians, whereas dark skin color seems of unique origin, reflecting the ancestral state in humans.
Link
mtDNA of Chad Basin populations
Annals of Human Genetics (OnlineEarly Articles)
A Bidirectional Corridor in the Sahel-Sudan Belt and the Distinctive Features of the Chad Basin Populations: A History Revealed by the Mitochondrial DNA Genome
V. Černý et al.
Summary
The Chad Basin was sparsely inhabited during the Stone Age, and its continual settlement began with the Holocene. The role played by Lake Chad in the history and migration patterns of Africa is still unclear. We studied the mitochondrial DNA (mtDNA) variability in 448 individuals from 12 ethnically and/or economically (agricultural/pastoral) different populations from Cameroon, Chad, Niger and Nigeria. The data indicate the importance of this region as a corridor connecting East and West Africa; however, this bidirectional flow of people in the Sahel-Sudan Belt did not erase features peculiar to the original Chad Basin populations. A new sub-clade, L3f2, is described, which together with L3e5 is most probably autochthonous in the Chad Basin. The phylogeography of these two sub-haplogroups seems to indicate prehistoric expansion events in the Chad Basin around 28,950 and 11,400 Y.B.P., respectively. The distribution of L3f2 is virtually restricted to the Chad Basin alone, and in particular to Chadic speaking populations, while L3e5 shows evidence for diffusion into North Africa at about 7,100 Y.B.P. The absence of L3f2 and L3e5 in African-Americans, and the limited number of L-haplotypes shared between the Chad Basin populations and African-Americans, indicate the low contribution of the Chad region to the Atlantic slave trade.
Link
A Bidirectional Corridor in the Sahel-Sudan Belt and the Distinctive Features of the Chad Basin Populations: A History Revealed by the Mitochondrial DNA Genome
V. Černý et al.
Summary
The Chad Basin was sparsely inhabited during the Stone Age, and its continual settlement began with the Holocene. The role played by Lake Chad in the history and migration patterns of Africa is still unclear. We studied the mitochondrial DNA (mtDNA) variability in 448 individuals from 12 ethnically and/or economically (agricultural/pastoral) different populations from Cameroon, Chad, Niger and Nigeria. The data indicate the importance of this region as a corridor connecting East and West Africa; however, this bidirectional flow of people in the Sahel-Sudan Belt did not erase features peculiar to the original Chad Basin populations. A new sub-clade, L3f2, is described, which together with L3e5 is most probably autochthonous in the Chad Basin. The phylogeography of these two sub-haplogroups seems to indicate prehistoric expansion events in the Chad Basin around 28,950 and 11,400 Y.B.P., respectively. The distribution of L3f2 is virtually restricted to the Chad Basin alone, and in particular to Chadic speaking populations, while L3e5 shows evidence for diffusion into North Africa at about 7,100 Y.B.P. The absence of L3f2 and L3e5 in African-Americans, and the limited number of L-haplotypes shared between the Chad Basin populations and African-Americans, indicate the low contribution of the Chad region to the Atlantic slave trade.
Link
January 16, 2007
ASPM and Microcephalin don't make people smarter
In Sep-05 a couple of papers showing that two brain genes, ASPM and Microcephalin had undergone positive selection in humans appeared in Science.
In May-06, another paper appeared that showed that Microcephalin and ASPM do not account for variability in brain size. This was a reasonable hypothesis to test, since these genes are implicated in microcephaly, a condition characterized by extremely small brains; it was thus natural to think that variation of these genes in normal subjects could also be correlated with their brain sizes; the data showed otherwise.
Now, a new study has appeared which studied the correlation of variants of these genes with IQ. As is well known, brain size only weakly correlates with IQ: in other words, by measuring how big a person's head is you get a very noisy estimate of how smart they are; other factors play a cumulatively bigger role. Since these genes are expressed in the brain, it might be possible that they worked their selective "magic" by conferring some advantage unrelated to brain size but related to IQ. However, even this is idea is also rejected by the data.
It remains to be seen what specific advantage -if any- the selected variants confer.
Hum Mol Genet. 2007 Jan 12; [Epub ahead of print]
The ongoing adaptive evolution of ASPM and Microcephalin is not explained by increased intelligence.
Mekel-Bobrov N et al.
Recent studies have made great strides towards identifying putative genetic events underlying the evolution of the human brain and its emergent cognitive capacities. One of the most intriguing findings is the recurrent identification of adaptive evolution in genes associated with primary microcephaly, a developmental disorder characterized by severe reduction in brain size and intelligence, reminiscent of the early hominid condition. This has led to the hypothesis that the adaptive evolution of these genes has contributed to the emergence of modern human cognition. As with other candidate loci, however, this hypothesis remains speculative due to the current lack of methodologies for characterizing the evolutionary function of these genes in humans. Two primary microcephaly genes, ASPM and Microcephalin, have been implicated not only in the adaptive evolution of the lineage leading to humans, but in ongoing selective sweeps in modern humans as well. The presence of both the putatively adaptive and neutral alleles at these loci provides a unique opportunity for using normal trait variation within humans to test the hypothesis that the recent selective sweeps are driven by an advantage in cognitive abilities. Here, we report a large-scale association study between the adaptive alleles of these genes and normal variation in several measures of IQ. Five independent samples were used, totaling 2,393 subjects, including both family-based and population-based datasets. Our overall findings do not support a detectable association between the recent adaptive evolution of either ASPM or Microcephalin and changes in IQ. As we enter the post-genomic era, with the number of candidate loci underlying human evolution growing rapidly, our findings highlight the importance of direct experimental validation in elucidating their evolutionary role in shaping the human phenotype.
Link
In May-06, another paper appeared that showed that Microcephalin and ASPM do not account for variability in brain size. This was a reasonable hypothesis to test, since these genes are implicated in microcephaly, a condition characterized by extremely small brains; it was thus natural to think that variation of these genes in normal subjects could also be correlated with their brain sizes; the data showed otherwise.
Now, a new study has appeared which studied the correlation of variants of these genes with IQ. As is well known, brain size only weakly correlates with IQ: in other words, by measuring how big a person's head is you get a very noisy estimate of how smart they are; other factors play a cumulatively bigger role. Since these genes are expressed in the brain, it might be possible that they worked their selective "magic" by conferring some advantage unrelated to brain size but related to IQ. However, even this is idea is also rejected by the data.
It remains to be seen what specific advantage -if any- the selected variants confer.
Hum Mol Genet. 2007 Jan 12; [Epub ahead of print]
The ongoing adaptive evolution of ASPM and Microcephalin is not explained by increased intelligence.
Mekel-Bobrov N et al.
Recent studies have made great strides towards identifying putative genetic events underlying the evolution of the human brain and its emergent cognitive capacities. One of the most intriguing findings is the recurrent identification of adaptive evolution in genes associated with primary microcephaly, a developmental disorder characterized by severe reduction in brain size and intelligence, reminiscent of the early hominid condition. This has led to the hypothesis that the adaptive evolution of these genes has contributed to the emergence of modern human cognition. As with other candidate loci, however, this hypothesis remains speculative due to the current lack of methodologies for characterizing the evolutionary function of these genes in humans. Two primary microcephaly genes, ASPM and Microcephalin, have been implicated not only in the adaptive evolution of the lineage leading to humans, but in ongoing selective sweeps in modern humans as well. The presence of both the putatively adaptive and neutral alleles at these loci provides a unique opportunity for using normal trait variation within humans to test the hypothesis that the recent selective sweeps are driven by an advantage in cognitive abilities. Here, we report a large-scale association study between the adaptive alleles of these genes and normal variation in several measures of IQ. Five independent samples were used, totaling 2,393 subjects, including both family-based and population-based datasets. Our overall findings do not support a detectable association between the recent adaptive evolution of either ASPM or Microcephalin and changes in IQ. As we enter the post-genomic era, with the number of candidate loci underlying human evolution growing rapidly, our findings highlight the importance of direct experimental validation in elucidating their evolutionary role in shaping the human phenotype.
Link
New study on Pestera cu Oase cranium from Romania

I don't see the study online yet [it is now online, see below], but here is the EurekAlert release:
Putting a face on the earliest modern EuropeansProc. Natl. Acad. Sci. USA, 10.1073/pnas.0610538104
The earliest modern humans in Europe were not completely "modern" and continued to evolve after they settled on the continent nearly 40,000 years ago, according to new research published this week in the Proceedings of the National Academy of Sciences (PNAS).
An international team of researchers, including Hélene Rougier, Ph.D., post-doctoral fellow, and Erik Trinkaus, Ph.D., professor of anthropology, at Washington University in St. Louis, has been studying a 35,000-year-old cranium discovered in the Pestera cu Oase, in western Romania. The fossil specimen is the earliest largely complete example of an early modern human skull known from Europe.
The modern humans emerged within eastern Africa some 150,000 years ago, spread temporarily into extreme southwest Asia and into southern Africa, and then through northern Africa into Europe around 40,000 years ago. This skull is from the first 5,000 years of the apparent occupation of Europe by modern humans.
The cranium has no expressly Neandertal traits, those of its immediate predecessors in Europe. Yet, its combination of modern and archaic features can be used to reinforce arguments for some degree of mixture of Neandertals and modern humans, inferences that have been made from other early modern European fossils. In addition to its large face and retreating forehead, it has the largest cheek teeth so far known for an otherwise anatomically modern human.
But Rougier and Trinkaus have emphasized that the Oase 2 cranium is particularly important in showing that the earliest modern humans in Europe were not completely "modern." "I think that what this find really shows is the ongoing nature of human evolution," said Trinkaus. "Technically, this skull is a modern human, but humans as we know them today have evolved considerably since then."
Pestera cu Oase 2 and the cranial morphology of early modern Europeans
Hélène Rougier et al.
Between 2003 and 2005, the Pestera cu Oase, Romania yielded a largely complete early modern human cranium, Oase 2, scattered on the surface of a Late Pleistocene hydraulically displaced bone bed containing principally the remains of Ursus spelaeus. Multiple lines of evidence indicate an age of {approx}40.5 thousand calendar years before the present ({approx}35 ka 14C B.P.). Morphological comparison of the adolescent Oase 2 cranium to relevant Late Pleistocene human samples documents a suite of derived modern human and/or non-Neandertal features, including absence of a supraorbital torus, subrectangular orbits, prominent canine fossae, narrow nasal aperture, level nasal floor, angled and anteriorly oriented zygomatic bones, a high neurocranium with prominent parietal bosses and marked sagittal parietal curvature, superiorly positioned temporal zygomatic root, vertical auditory porous, laterally bulbous mastoid processes, superiorly positioned posterior semicircular canal, absence of a nuchal torus and a suprainiac fossa, and a small occipital bun. However, these features are associated with an exceptionally flat frontal arc, a moderately large juxtamastoid eminence, extremely large molars that become progressively larger distally, complex occlusal morphology of the upper third molar, and relatively anteriorly positioned zygomatic arches. Moreover, the featureless occipital region and small mastoid process are at variance with the large facial skeleton and dentition. This unusual mosaic in Oase 2, some of which is paralleled in the Oase 1 mandible, indicates both complex population dynamics as modern humans dispersed into Europe and significant ongoing human evolution once modern humans were established within Europe.
Link
January 11, 2007
New Kostenki and South African finds in Science magazine (earliest occupation of Eurasia by modern humans)
I didn't see the papers online yet [they are now online], but from EurekAlert:
It also stresses the idea that contrary to popular treatments such as Spencer Wells' Journey of Man, the appearance of African hunter-gatherers such as the Khoi-San should not be used as a model of what mankind's earliest African ancestors looked like.
Late Pleistocene Human Skull from Hofmeyr, South Africa, and Modern Human Origins
F. E. Grine et al.
The lack of Late Pleistocene human fossils from sub-Saharan Africa has limited paleontological testing of competing models of recent human evolution. We have dated a skull from Hofmeyr, South Africa, to 36.2 ± 3.3 thousand years ago through a combination of optically stimulated luminescence and uranium-series dating methods. The skull is morphologically modern overall but displays some archaic features. Its strongest morphometric affinities are with Upper Paleolithic (UP) Eurasians rather than recent, geographically proximate people. The Hofmeyr cranium is consistent with the hypothesis that UP Eurasians descended from a population that emigrated from sub-Saharan Africa in the Late Pleistocene.
Link
Early Upper Paleolithic in Eastern Europe and Implications for the Dispersal of Modern Humans
M. V. Anikovich et al.
Radiocarbon and optically stimulated luminescence dating and magnetic stratigraphy indicate Upper Paleolithic occupation—probably representing modern humans—at archaeological sites on the Don River in Russia 45,000 to 42,000 years ago. The oldest levels at Kostenki underlie a volcanic ash horizon identified as the Campanian Ignimbrite Y5 tephra that is dated elsewhere to about 40,000 years ago. The occupation layers contain bone and ivory artifacts, including possible figurative art, and fossil shells imported more than 500 kilometers. Thus, modern humans appeared on the central plain of Eastern Europe as early as anywhere else in northern Eurasia.
Link
The excavation took place at Kostenki, a group of more than 20 sites along the Don River that have been under study for many decades. Kostenki previously has yielded anatomically modern human bones and artifacts dating between 30,000 and 40,000 years old, including the oldest firmly dated bone and ivory needles with eyelets that indicate the early inhabitants were tailoring animal furs to help them survive the harsh climate.From the New York Times:
An international team of researchers reported today that the age of the South African skull, which they dated at about 36,000 years old, coincided with the age of and closely resembled the skulls of humans who were then living in Europe and the far eastern parts of Asia, even Australia.The discovery of these resemblances lends some support to the idea that I have expressed previously that only a subset of modern humans (which I have called Afrasians) was responsible for the colonization of Eurasia. At the time frame in question, these resemblances would have been marked, since (i) intermixture with pre-existing populations (such as "Paleoafricans" or pre-moderns in Europe) did not have time to take place yet, and (ii) racial differentiation had only just begun. This paragraph says it all:
Because the Bushmen are well represented in the more recent archaeological record, Dr. Harvati said, they were expected to bear a close resemblance to the Hofmeyr skull. Instead, the skull was found to be quite distinct from all recent Africans, including the Bushmen, she said, and it has “a very close affinity” with fossil specimens of Europeans living in the Upper Paleolithic, the period best known for advanced stone tools and cave art.The dissimilarity to recent Africans of the Hofmeyr skull can be easily explained if it is understood that recent Africans are not only descended from the "Afrasians" that I have spoken of, but also from the older "Paleofricans" whose existence can be inferred both by genetics (human mtDNA and Y-chromosomes are older than the ~40kya mark) and paleoanthropology (modern humans such as Omo and Herto are 100-200kya old).
“Much to my amazement,” Dr. Grine said in an interview, “the skull linked very closely with those from Europe at the time and not with South African remains 15,000 years on.”
It also stresses the idea that contrary to popular treatments such as Spencer Wells' Journey of Man, the appearance of African hunter-gatherers such as the Khoi-San should not be used as a model of what mankind's earliest African ancestors looked like.
Late Pleistocene Human Skull from Hofmeyr, South Africa, and Modern Human Origins
F. E. Grine et al.
The lack of Late Pleistocene human fossils from sub-Saharan Africa has limited paleontological testing of competing models of recent human evolution. We have dated a skull from Hofmeyr, South Africa, to 36.2 ± 3.3 thousand years ago through a combination of optically stimulated luminescence and uranium-series dating methods. The skull is morphologically modern overall but displays some archaic features. Its strongest morphometric affinities are with Upper Paleolithic (UP) Eurasians rather than recent, geographically proximate people. The Hofmeyr cranium is consistent with the hypothesis that UP Eurasians descended from a population that emigrated from sub-Saharan Africa in the Late Pleistocene.
Link
Early Upper Paleolithic in Eastern Europe and Implications for the Dispersal of Modern Humans
M. V. Anikovich et al.
Radiocarbon and optically stimulated luminescence dating and magnetic stratigraphy indicate Upper Paleolithic occupation—probably representing modern humans—at archaeological sites on the Don River in Russia 45,000 to 42,000 years ago. The oldest levels at Kostenki underlie a volcanic ash horizon identified as the Campanian Ignimbrite Y5 tephra that is dated elsewhere to about 40,000 years ago. The occupation layers contain bone and ivory artifacts, including possible figurative art, and fossil shells imported more than 500 kilometers. Thus, modern humans appeared on the central plain of Eastern Europe as early as anywhere else in northern Eurasia.
Link
Age-related immigration to imperial Rome
Am J Phys Anthropol. 2007 Jan 4; [Epub ahead of print]
Isotopic evidence for age-related immigration to imperial Rome
Browse TL et al.
Oxygen stable isotope ratios (delta(18)O) have been determined in carbonate in paired first and third molar teeth from individuals (N = 61) who lived in the town of Portus Romae ("Portus") and who were buried in the necropolis of Isola Sacra (First to Third centuries AD) near Rome, Italy. We compare these analyses with data for deciduous teeth of modern Roman children. Approximately one-third of the archaeological sample has first molar (M1) values outside the modern range, implying a large rate of population turnover at that time, consistent with historical data. Delta (18)O(ap) values suggest that a group within the sample migrated to the area before the third molar (M3) crown had completely formed (i.e., between 10 and 17.5 years of age). This is the first quantitative assessment of population mobility in Classical antiquity. This study demonstrates that migration was not limited to predominantly single adult males, as suggested by historical sources, but rather a complex phenomenon involving families. We hypothesize that migrants most likely came from higher elevations to the East and North of Rome. One individual with a higher delta(18)O value may have come (as a child) from an area isotopically similar to North Africa.
Link
Isotopic evidence for age-related immigration to imperial Rome
Browse TL et al.
Oxygen stable isotope ratios (delta(18)O) have been determined in carbonate in paired first and third molar teeth from individuals (N = 61) who lived in the town of Portus Romae ("Portus") and who were buried in the necropolis of Isola Sacra (First to Third centuries AD) near Rome, Italy. We compare these analyses with data for deciduous teeth of modern Roman children. Approximately one-third of the archaeological sample has first molar (M1) values outside the modern range, implying a large rate of population turnover at that time, consistent with historical data. Delta (18)O(ap) values suggest that a group within the sample migrated to the area before the third molar (M3) crown had completely formed (i.e., between 10 and 17.5 years of age). This is the first quantitative assessment of population mobility in Classical antiquity. This study demonstrates that migration was not limited to predominantly single adult males, as suggested by historical sources, but rather a complex phenomenon involving families. We hypothesize that migrants most likely came from higher elevations to the East and North of Rome. One individual with a higher delta(18)O value may have come (as a child) from an area isotopically similar to North Africa.
Link
January 10, 2007
Reduced Y chromosome diversity of Central Asian pastoral populations
Curr Biol. 2007 Jan 9;17(1):43-8.
From social to genetic structures in central Asia.
Chaix R. et al.
Pastoral and farmer populations, who have coexisted in Central Asia since the fourth millennium B.C. , present not only different lifestyles and means of subsistence but also various types of social organization. Pastoral populations are organized into so-called descent groups (tribes, clans, and lineages) and practice exogamous marriages (a man chooses a bride in a different lineage or clan). In Central Asia, these descent groups are patrilineal: The children are systematically affiliated with the descent groups of the father. By contrast, farmer populations are organized into families (extended or nuclear) and often establish endogamous marriages with cousins . This study aims at better understanding the impact of these differences in lifestyle and social organization on the shaping of genetic diversity. We show that pastoral populations exhibit a substantial loss of Y chromosome diversity in comparison to farmers but that no such a difference is observed at the mitochondrial-DNA level. Our analyses indicate that the dynamics of patrilineal descent groups, which implies different male and female sociodemographic histories, is responsible for these sexually-asymmetric genetic patterns. This molecular signature of the pastoral social organization disappears over a few centuries only after conversion to an agricultural way of life.
Link
From social to genetic structures in central Asia.
Chaix R. et al.
Pastoral and farmer populations, who have coexisted in Central Asia since the fourth millennium B.C. , present not only different lifestyles and means of subsistence but also various types of social organization. Pastoral populations are organized into so-called descent groups (tribes, clans, and lineages) and practice exogamous marriages (a man chooses a bride in a different lineage or clan). In Central Asia, these descent groups are patrilineal: The children are systematically affiliated with the descent groups of the father. By contrast, farmer populations are organized into families (extended or nuclear) and often establish endogamous marriages with cousins . This study aims at better understanding the impact of these differences in lifestyle and social organization on the shaping of genetic diversity. We show that pastoral populations exhibit a substantial loss of Y chromosome diversity in comparison to farmers but that no such a difference is observed at the mitochondrial-DNA level. Our analyses indicate that the dynamics of patrilineal descent groups, which implies different male and female sociodemographic histories, is responsible for these sexually-asymmetric genetic patterns. This molecular signature of the pastoral social organization disappears over a few centuries only after conversion to an agricultural way of life.
Link
January 07, 2007
Linguistic and Genetic Relationships of Central and South Americans
Am J Phys Anthropol. 2007 Jan 4; [Epub ahead of print]
A formal test of linguistic and genetic coevolution in native Central and South America.
Hunley KL et al.
This paper investigates a mechanism of linguistic and genetic coevolution in Native Central and South America. This mechanism proposes that a process of population fissions, expansions into new territories, and isolation of ancestral and descendant groups will produce congruent language and gene trees. To evaluate this population fissions mechanism, we collected published mtDNA sequences for 1,381 individuals from 17 Native Central and South American populations. We then tested the hypothesis that three well-known language classifications also represented the genetic structure of these populations. We rejected the hypothesis for each language classification. Our tests revealed linguistic and genetic correspondence in several shallow branches common to each classification, but no linguistic and genetic correspondence in the deeper branches contained in two of the language classifications. We discuss the possible causes for the lack of congruence between linguistic and genetic structure in the region, and describe alternative mechanisms of linguistic and genetic correspondence and their predictions.
Link
A formal test of linguistic and genetic coevolution in native Central and South America.
Hunley KL et al.
This paper investigates a mechanism of linguistic and genetic coevolution in Native Central and South America. This mechanism proposes that a process of population fissions, expansions into new territories, and isolation of ancestral and descendant groups will produce congruent language and gene trees. To evaluate this population fissions mechanism, we collected published mtDNA sequences for 1,381 individuals from 17 Native Central and South American populations. We then tested the hypothesis that three well-known language classifications also represented the genetic structure of these populations. We rejected the hypothesis for each language classification. Our tests revealed linguistic and genetic correspondence in several shallow branches common to each classification, but no linguistic and genetic correspondence in the deeper branches contained in two of the language classifications. We discuss the possible causes for the lack of congruence between linguistic and genetic structure in the region, and describe alternative mechanisms of linguistic and genetic correspondence and their predictions.
Link
DNA of the extinct Beothuk
Am J Phys Anthropol. 2007 Jan 4; [Epub ahead of print]
A preliminary analysis of the DNA and diet of the extinct Beothuk: A systematic approach to ancient human DNA.
Kuch M et al.
We have used a systematic protocol for extracting, quantitating, sexing and validating ancient human mitochondrial and nuclear DNA of one male and one female Beothuk, a Native American population from Newfoundland, which became extinct approximately 180 years ago. They carried mtDNA haplotypes, which fall within haplogroups X and C, consistent with Northeastern Native populations today. In addition we have sexed the male using a novel-sexing assay and confirmed the authenticity of his Y chromosome with the presence of the Native American specific Y-QM3 single nucleotide polymorphism (SNP). This is the first ancient nuclear SNP typed from a Native population in the Americas. In addition, using the same teeth we conducted a stable isotopes analysis of collagen and dentine to show that both individuals relied on marine sources (fresh and salt water fish, seals) with no hierarchy seen between them, and that their water sources were pooled or stored water. Both mtDNA sequence data and Y SNP data hint at possible gene flow or a common ancestral population for both the Beothuk and the current day Mikmaq, but more importantly the data do not lend credence to the proposed idea that the Beothuk (specifically, Nonosabasut) were of admixed (European-Native American) descent. We also analyzed patterns of DNA damage in the clones of authentic mtDNA sequences; there is no tendency for DNA damage to occur preferentially at previously defined mutational hotspots, suggesting that such mutational hotspots are not hypervariable because they are more prone to damage.
Link
A preliminary analysis of the DNA and diet of the extinct Beothuk: A systematic approach to ancient human DNA.
Kuch M et al.
We have used a systematic protocol for extracting, quantitating, sexing and validating ancient human mitochondrial and nuclear DNA of one male and one female Beothuk, a Native American population from Newfoundland, which became extinct approximately 180 years ago. They carried mtDNA haplotypes, which fall within haplogroups X and C, consistent with Northeastern Native populations today. In addition we have sexed the male using a novel-sexing assay and confirmed the authenticity of his Y chromosome with the presence of the Native American specific Y-QM3 single nucleotide polymorphism (SNP). This is the first ancient nuclear SNP typed from a Native population in the Americas. In addition, using the same teeth we conducted a stable isotopes analysis of collagen and dentine to show that both individuals relied on marine sources (fresh and salt water fish, seals) with no hierarchy seen between them, and that their water sources were pooled or stored water. Both mtDNA sequence data and Y SNP data hint at possible gene flow or a common ancestral population for both the Beothuk and the current day Mikmaq, but more importantly the data do not lend credence to the proposed idea that the Beothuk (specifically, Nonosabasut) were of admixed (European-Native American) descent. We also analyzed patterns of DNA damage in the clones of authentic mtDNA sequences; there is no tendency for DNA damage to occur preferentially at previously defined mutational hotspots, suggesting that such mutational hotspots are not hypervariable because they are more prone to damage.
Link
January 05, 2007
Facial masculinity
From LiveScience:
Personal Relationships
Volume 13 Issue 4 Page 451 - December 2006
Male facial masculinity influences attributions of personality and reproductive strategy
DANIEL J. KRUGER
Abstract
Facial masculinity may be used as a cue in female mate choice, as it reflects the success of the male genotype in its developmental environment. Women may maximize reproductive success by using a conditional strategy favoring highly masculine facial features for short-term relationships and feminized facial features in men for long-term relationships. Three studies examine reactions to masculinized and feminized male facial composites. Properties of the original composite image affect ratings of critical attributes and the magnitude of the differences in ratings between versions undergoing identical processes of geometric manipulation (Study 1). Both men and women attribute personality, behavior, and mating strategies consistent with predictions derived from the good genes and mating trade-off hypotheses (Study 2). Participants accurately grouped behavioral tendencies related to high mating effort/risky strategies and high parenting effort/risk adverse strategies and associated mating effort more so with masculinized faces and parenting effort more so with feminized faces (Study 3). These results indicate that male facial masculinity serves as a visual cue for inferring personality and reproductive strategy.
Link
In the study, 854 male and female subjects viewed a series of male head shots that had been digitally altered to exaggerate or minimize masculine traits. The participants then answered questions about how they expected the men in the photos to behave.
Overwhelmingly, participants said those with more masculine features were likely to be risky and competitive and also more apt to fight, challenge bosses, cheat on spouses and put less effort into parenting. Those with more feminine faces were seen as good parents and husbands, hard workers and emotionally supportive mates [compare examples].
Despite all the negative attributes, when asked who they would choose for a short-term relationship, women still selected the more masculine looking men. Brad and George then would be picks for a brief romance, if not the long haul.
Personal Relationships
Volume 13 Issue 4 Page 451 - December 2006
Male facial masculinity influences attributions of personality and reproductive strategy
DANIEL J. KRUGER
Abstract
Facial masculinity may be used as a cue in female mate choice, as it reflects the success of the male genotype in its developmental environment. Women may maximize reproductive success by using a conditional strategy favoring highly masculine facial features for short-term relationships and feminized facial features in men for long-term relationships. Three studies examine reactions to masculinized and feminized male facial composites. Properties of the original composite image affect ratings of critical attributes and the magnitude of the differences in ratings between versions undergoing identical processes of geometric manipulation (Study 1). Both men and women attribute personality, behavior, and mating strategies consistent with predictions derived from the good genes and mating trade-off hypotheses (Study 2). Participants accurately grouped behavioral tendencies related to high mating effort/risky strategies and high parenting effort/risk adverse strategies and associated mating effort more so with masculinized faces and parenting effort more so with feminized faces (Study 3). These results indicate that male facial masculinity serves as a visual cue for inferring personality and reproductive strategy.
Link
Forbidden DNA sequences
Via the New Scientist:
Could there be forbidden sequences in the genome - ones so harmful that they are not compatible with life? One group of researchers thinks so. Unlike most genome sequencing projects which set out to search for genes that are conserved within and between species, their goal is to identify "primes": DNA sequences and chains of amino acids so dangerous to life that they do not exist.In a back of the envelope calculation, a particular sequence of 11 nucleotides has a 4-11 chance of occurring, but there are around 3 x 109 11-nucleotide sequences in the human genome. Thus, it is very unlikely that such a sequence will not occur by accident in a human. Of course there are repetitive elements in the human genome, as well as functional elements that have been conserved intact across our species and even across millions of years of evolution that separate us from other taxa. Thus, our genomes are not 3-billion long random strings of CAGT. However, the principle is valid that short sequences have a higher chance of occurring by accident in longer genomes. Thus, the fact that some sequences may not occur at all is fairly good evidence that they "do something bad" and hence nature avoids them. From the article:
...
To do this, Hampikian and his colleage Tim Anderson, also at Boise, have developed software that calculates all the possible sequences of nucleotides - the "letters" of DNA - up to a certain length, and then scans sequence databases such as the US National Institutes of Health's Genbank to identify the smallest sequences that aren't present. Those that don't occur in one species but do in others are termed "nullomers", while those that aren't found in any species are termed primes.
Hampikian's team is deliberately searching for the shortest absent sequences in order to minimise the possibility that absent sequences are missing simply due to chance. So far they have found 86 sequences of 11 nucleotides long that have never been reported in humans.
Whether these sequences have any biological significance in living organisms is not yet known - the next step is to test 20 of the peptoprimes in bacteria and human cells to see whether they have any effect such as causing death or provoking an immune reaction.
Subscribe to:
Comments (Atom)
