In some cases, small bands of potentially as few as 20 to 30 people could have been moving over very large areas, over the whole of Europe as a single territory, according to Professor Ron Pinhasi, principal investigator on the EU-funded ADNABIOARC project.The last couple of statements are interesting because they hint at post-glacial recolonization of Europe after the Ice Age. So far, we are in the dark about what happened in Europe between the time of Kostenki and 8kya. Hopefully another interesting study is on its way to throw some light into the lattter part of this time interval.
This demographic model is based on new evidence that suggests populations were much smaller than is generally thought to be a stable size for healthy reproduction, usually around 500 people. Such small groupings may have led to reduced fitness and even extinctions.
‘As an archaeologist and anthropologist, I was quite shocked to see how limited, how small the population numbers were. You know, shockingly small,’ said Prof. Pinhasi, based at University College Dublin, Ireland.
...
Prof. Pinhasi’s team has found that the genomes sequenced from hunter-gatherers from Hungary and Switzerland between 14 000 to 7 500 years ago are very close to specimens from Denmark or Sweden from the same period.
These findings suggest that genetic diversity between inhabitants of most of western and central Europe after the ice age was very limited, indicating a major demographic bottleneck triggered by human isolation and extinction during the ice age.
‘We’re starting to be able to reconstruct the actual dynamics of migrations and colonisation of the continent by modern humans and that’s never been done before the genomic era,’ explained Prof. Pinhasi.
He believes that early humans crossed the continent in small groups that were cut off while the ice was at its peak, then successively dispersed and regrouped over thousands of years, with dwindling northern populations invigorated by humans arriving from the south, where the climate was better.
‘You see a real reduction in population numbers and diversity, so you see the few lineages that probably split or separated before the ice age, and then stayed isolated during the ice age,’ he said. ‘Some time after the ice age, they kind of re-emerge, or disperse, and get together, as we see new contributions to European lineages from Asia and in particular the Near East.’
March 30, 2015
Ice age Europeans on the brink of extinction
Ice-age Europeans roamed in small bands of fewer than 30, on brink of extinction (Horizon magazine)
March 28, 2015
Afanasievo, Okunev, Andronovo, Sintashta DNA?
A reader alerts me to this article in Russian, but you can use Google Translate to get the gist of it. Some interesting bits (note that "pit"=Yamna):
I can not ignore the question I now have is particularly exciting - the origin of the Indo-Europeans. Community Indo-Europeists animatedly discussing just appeared as a preprint work of David Raika and his colleagues discovered by studying the genomes of people Neolithic and Bronze Age that a decisive influence on the genetic landscape of Europe has had a migration of people pit culture to the north and west in the middle of the III millennium. BC .e. As a result, according to geneticists, there was a population associated with the Corded Ware culture, and from it are the origin of the later Indo-European. By the same conclusions about the same time came the other team's leading geneticists led by Eske Villerslevom.I hope more details will appear soon on what promises to be a very interesting new study. The author seems to be referring to his theory of a relationship between Okunev and Amerindians, and I'm wondering if this is simply "Ancient North Eurasian" ancestry or an even more specific link. Any Russian readers who can dig up more information are invited to post in the comments.
...
A steppe, we thought had long been a special world, and differs sharply from the Middle East, and from the European. Migration from there - so it seemed - were mainly directed not to the west and to the east, along the steppes, in the direction of Central Asia, which the ancient Indo-Europeans, Afanasiev media culture (descendants of the people of the pit culture or their ancestors steppe) reached no later turn IV- III millennium BC. It is now confirmed and the group Villersleva.
...
By the way, it also happens that paleoanthropologists prompted geneticists way of research - and turned out to be right. As it happens, for example, with native Okunevskaya culture of South Siberia. When 20 years ago, we found that craniologically (by a combination of traditional measurement and we proposed new informative features of the structure of the cranial sutures and holes) okunevtsy - "cousins" of American Indians, few believed us. Firstly, in okunevtsah ever seen Caucasoid-Mongoloid Métis like the Kazakhs, and secondly, the ancestors of the Indians withdrew from Siberia to the New World at least 10 thousand. Before the Yenisey there Okunevskaya culture.
Eske Willerslev Now and his colleagues have fully confirmed our conclusion. They confirmed the close relationship between the carriers and the pit Afanasiev cultures and migration ancestors sintashtintsev and Andronov from Europe in the Urals and further to Siberia - but this is already a long time, few archaeologists and anthropologists doubted.
March 25, 2015
Icelanders galore
A set of four papers in Nature Genetics today. All open access. Of interest from the Y-chromosome paper:
Nature Genetics (2015) doi:10.1038/ng.3247
Large-scale whole-genome sequencing of the Icelandic population
Daniel F Gudbjartsson et al.
Here we describe the insights gained from sequencing the whole genomes of 2,636 Icelanders to a median depth of 20×. We found 20 million SNPs and 1.5 million insertions-deletions (indels). We describe the density and frequency spectra of sequence variants in relation to their functional annotation, gene position, pathway and conservation score. We demonstrate an excess of homozygosity and rare protein-coding variants in Iceland. We imputed these variants into 104,220 individuals down to a minor allele frequency of 0.1% and found a recessive frameshift mutation in MYL4 that causes early-onset atrial fibrillation, several mutations in ABCB4 that increase risk of liver diseases and an intronic variant in GNAS associating with increased thyroid-stimulating hormone levels when maternally inherited. These data provide a study design that can be used to determine how variation in the sequence of the human genome gives rise to human diversity.
Link
Nature Genetics (2015) doi:10.1038/ng.3171
The Y-chromosome point mutation rate in humans
Agnar Helgason et al.
Mutations are the fundamental source of biological variation, and their rate is a crucial parameter for evolutionary and medical studies. Here we used whole-genome sequence data from 753 Icelandic males, grouped into 274 patrilines, to estimate the point mutation rate for 21.3 Mb of male-specific Y chromosome (MSY) sequence, on the basis of 1,365 meioses (47,123 years). The combined mutation rate for 15.2 Mb of X-degenerate (XDG), X-transposed (XTR) and ampliconic excluding palindromes (rAMP) sequence was 8.71 × 10−10 mutations per position per year (PPPY). We observed a lower rate (P = 0.04) of 7.37 × 10−10 PPPY for 6.1 Mb of sequence from palindromes (PAL), which was not statistically different from the rate of 7.2 × 10−10 PPPY for paternally transmitted autosomes1. We postulate that the difference between PAL and the other MSY regions may provide an indication of the rate at which nascent autosomal and PAL de novo mutations are repaired as a result of gene conversion.
Link
Nature Genetics (2015) doi:10.1038/ng.3246
Loss-of-function variants in ABCA7 confer risk of Alzheimer's disease
Stacy Steinberg et al.
We conducted a search for rare, functional variants altering susceptibility to Alzheimer's disease that exploited knowledge of common variants associated with the same disease. We found that loss-of-function variants in ABCA7 confer risk of Alzheimer's disease in Icelanders (odds ratio (OR) = 2.12, P = 2.2 × 10−13) and discovered that the association replicated in study groups from Europe and the United States (combined OR = 2.03, P = 6.8 × 10−15).
Link
Nature Genetics (2015) doi:10.1038/ng.3243
Identification of a large set of rare complete human knockouts
Patrick Sulem et al.
Loss-of-function mutations cause many mendelian diseases. Here we aimed to create a catalog of autosomal genes that are completely knocked out in humans by rare loss-of-function mutations. We sequenced the whole genomes of 2,636 Icelanders and imputed the sequence variants identified in this set into 101,584 additional chip-genotyped and phased Icelanders. We found a total of 6,795 autosomal loss-of-function SNPs and indels in 4,924 genes. Of the genotyped Icelanders, 7.7% are homozygotes or compound heterozygotes for loss-of-function mutations with a minor allele frequency (MAF) below 2% in 1,171 genes (complete knockouts). Genes that are highly expressed in the brain are less often completely knocked out than other genes. Homozygous loss-of-function offspring of two heterozygous parents occurred less frequently than expected (deficit of 136 per 10,000 transmissions for variants with MAF less than 2%, 95% confidence interval (CI) = 10–261).
Link
When this rate was applied to estimate the TMRCA between two Y chromosomes that encompass the oldest known patrilineal bifurcation between any humans (representing haplogroups A00 and A0, with 75 derived mutational differences in 180 kb of XDG sequence)19, we obtained a maximum-likelihood estimate21 of 239,000 years ago and a 95% CI of 188,000–296,000 years ago (174,000–321,000 years ago when incorporating the 95% CI of our mutation rate).This seems similar to the 254kya estimated by Karmin et al.
Nature Genetics (2015) doi:10.1038/ng.3247
Large-scale whole-genome sequencing of the Icelandic population
Daniel F Gudbjartsson et al.
Here we describe the insights gained from sequencing the whole genomes of 2,636 Icelanders to a median depth of 20×. We found 20 million SNPs and 1.5 million insertions-deletions (indels). We describe the density and frequency spectra of sequence variants in relation to their functional annotation, gene position, pathway and conservation score. We demonstrate an excess of homozygosity and rare protein-coding variants in Iceland. We imputed these variants into 104,220 individuals down to a minor allele frequency of 0.1% and found a recessive frameshift mutation in MYL4 that causes early-onset atrial fibrillation, several mutations in ABCB4 that increase risk of liver diseases and an intronic variant in GNAS associating with increased thyroid-stimulating hormone levels when maternally inherited. These data provide a study design that can be used to determine how variation in the sequence of the human genome gives rise to human diversity.
Link
Nature Genetics (2015) doi:10.1038/ng.3171
The Y-chromosome point mutation rate in humans
Agnar Helgason et al.
Mutations are the fundamental source of biological variation, and their rate is a crucial parameter for evolutionary and medical studies. Here we used whole-genome sequence data from 753 Icelandic males, grouped into 274 patrilines, to estimate the point mutation rate for 21.3 Mb of male-specific Y chromosome (MSY) sequence, on the basis of 1,365 meioses (47,123 years). The combined mutation rate for 15.2 Mb of X-degenerate (XDG), X-transposed (XTR) and ampliconic excluding palindromes (rAMP) sequence was 8.71 × 10−10 mutations per position per year (PPPY). We observed a lower rate (P = 0.04) of 7.37 × 10−10 PPPY for 6.1 Mb of sequence from palindromes (PAL), which was not statistically different from the rate of 7.2 × 10−10 PPPY for paternally transmitted autosomes1. We postulate that the difference between PAL and the other MSY regions may provide an indication of the rate at which nascent autosomal and PAL de novo mutations are repaired as a result of gene conversion.
Link
Nature Genetics (2015) doi:10.1038/ng.3246
Loss-of-function variants in ABCA7 confer risk of Alzheimer's disease
Stacy Steinberg et al.
We conducted a search for rare, functional variants altering susceptibility to Alzheimer's disease that exploited knowledge of common variants associated with the same disease. We found that loss-of-function variants in ABCA7 confer risk of Alzheimer's disease in Icelanders (odds ratio (OR) = 2.12, P = 2.2 × 10−13) and discovered that the association replicated in study groups from Europe and the United States (combined OR = 2.03, P = 6.8 × 10−15).
Link
Nature Genetics (2015) doi:10.1038/ng.3243
Identification of a large set of rare complete human knockouts
Patrick Sulem et al.
Loss-of-function mutations cause many mendelian diseases. Here we aimed to create a catalog of autosomal genes that are completely knocked out in humans by rare loss-of-function mutations. We sequenced the whole genomes of 2,636 Icelanders and imputed the sequence variants identified in this set into 101,584 additional chip-genotyped and phased Icelanders. We found a total of 6,795 autosomal loss-of-function SNPs and indels in 4,924 genes. Of the genotyped Icelanders, 7.7% are homozygotes or compound heterozygotes for loss-of-function mutations with a minor allele frequency (MAF) below 2% in 1,171 genes (complete knockouts). Genes that are highly expressed in the brain are less often completely knocked out than other genes. Homozygous loss-of-function offspring of two heterozygous parents occurred less frequently than expected (deficit of 136 per 10,000 transmissions for variants with MAF less than 2%, 95% confidence interval (CI) = 10–261).
Link
March 21, 2015
Ancient mtDNA from cis-Baikal area
Russian Journal of Genetics: Applied Research
January 2015, Volume 5, Issue 1, pp 26-32
Mitochondrial DNA diversity in the gene pool of the Neolithic and Early Bronze Age Cisbaikalian human population R. O. Trapezov, A. S. Pilipenko, V. I. Molodin
This paper presents the results of a study of a mitochondrial DNA sample (N = 15) from the remains of representatives of the Neolithic and Early Bronze Age (VI–III millennia BC) Cisbaikalian human population. It was found that the mitochondrial gene pool of the ancient population under study contains lineages of East Eurasian haplogroups D, G2a C, Z, and F1b. The results of the comparative analysis of the group under study with ancient and modern Eurasian populations suggest that the development of autochtonous East Eurasian genetic components was the main mechanism of the formation of the population of the Baikal region. Genetic contacts with populations of neighboring regions of Central Asia also contributed to the formation of the gene pool of the Cisbaikalian population.
Link
Mitochondrial DNA diversity in the gene pool of the Neolithic and Early Bronze Age Cisbaikalian human population R. O. Trapezov, A. S. Pilipenko, V. I. Molodin
This paper presents the results of a study of a mitochondrial DNA sample (N = 15) from the remains of representatives of the Neolithic and Early Bronze Age (VI–III millennia BC) Cisbaikalian human population. It was found that the mitochondrial gene pool of the ancient population under study contains lineages of East Eurasian haplogroups D, G2a C, Z, and F1b. The results of the comparative analysis of the group under study with ancient and modern Eurasian populations suggest that the development of autochtonous East Eurasian genetic components was the main mechanism of the formation of the population of the Baikal region. Genetic contacts with populations of neighboring regions of Central Asia also contributed to the formation of the gene pool of the Cisbaikalian population.
Link
March 20, 2015
Campanian Ignibrite and Neandertal demise
Geology doi:10.1130/G36514.1
Campanian Ignimbrite volcanism, climate, and the final decline of the Neanderthals
Benjamin A. Black1, Ryan R. Neely2,3,4 and Michael Manga1
The eruption of the Campanian Ignimbrite at ca. 40 ka coincided with the final decline of Neanderthals in Europe. Environmental stress associated with the eruption of the Campanian Ignimbrite has been invoked as a potential driver for this extinction as well as broader upheaval in Paleolithic societies. To test the climatic importance of the Campanian eruption, we used a three-dimensional sectional aerosol model to simulate the global aerosol cloud after release of 50 Tg and 200 Tg SO2. We coupled aerosol properties to a comprehensive earth system model under last glacial conditions. We find that peak cooling and acid deposition lasted one to two years and that the most intense cooling sidestepped hominin population centers in Western Europe. We conclude that the environmental effects of the Campanian Ignimbrite eruption alone were insufficient to explain the ultimate demise of Neanderthals in Europe. Nonetheless, significant volcanic cooling during the years immediately following the eruption could have impacted the viability of already precarious populations and influenced many aspects of daily life for Neanderthals and anatomically modern humans.
Link
Campanian Ignimbrite volcanism, climate, and the final decline of the Neanderthals
Benjamin A. Black1, Ryan R. Neely2,3,4 and Michael Manga1
The eruption of the Campanian Ignimbrite at ca. 40 ka coincided with the final decline of Neanderthals in Europe. Environmental stress associated with the eruption of the Campanian Ignimbrite has been invoked as a potential driver for this extinction as well as broader upheaval in Paleolithic societies. To test the climatic importance of the Campanian eruption, we used a three-dimensional sectional aerosol model to simulate the global aerosol cloud after release of 50 Tg and 200 Tg SO2. We coupled aerosol properties to a comprehensive earth system model under last glacial conditions. We find that peak cooling and acid deposition lasted one to two years and that the most intense cooling sidestepped hominin population centers in Western Europe. We conclude that the environmental effects of the Campanian Ignimbrite eruption alone were insufficient to explain the ultimate demise of Neanderthals in Europe. Nonetheless, significant volcanic cooling during the years immediately following the eruption could have impacted the viability of already precarious populations and influenced many aspects of daily life for Neanderthals and anatomically modern humans.
Link
March 18, 2015
British origins (Leslie et al. 2015)
The long-awaited paper on the People of the British Isles has just appeared in Nature. I will update this entry with more information.
UPDATE:
The authors write:
Most of the clusters are geographical, but some span different regions (e.g., the "yellow circle" cluster). The elephant in the room is the "red square" cluster which spans Central/South England. The authors write:
The authors draw conclusions on several historical episodes of British history. The big one is the extent of Anglo-Saxon ancestry:
Nature 519, 309–314 (19 March 2015) doi:10.1038/nature14230
The fine-scale genetic structure of the British population
Stephen Leslie et al.
Fine-scale genetic variation between human populations is interesting as a signature of historical demographic events and because of its potential for confounding disease studies. We use haplotype-based statistical methods to analyse genome-wide single nucleotide polymorphism (SNP) data from a carefully chosen geographically diverse sample of 2,039 individuals from the United Kingdom. This reveals a rich and detailed pattern of genetic differentiation with remarkable concordance between genetic clusters and geography. The regional genetic differentiation and differing patterns of shared ancestry with 6,209 individuals from across Europe carry clear signals of historical demographic events. We estimate the genetic contribution to southeastern England from Anglo-Saxon migrations to be under half, and identify the regions not carrying genetic material from these migrations. We suggest significant pre-Roman but post-Mesolithic movement into southeastern England from continental Europe, and show that in non-Saxon parts of the United Kingdom, there exist genetically differentiated subgroups rather than a general ‘Celtic’ population.
Link
UPDATE:
The authors write:
Consistent with earlier studies of the UK, population structure within the PoBI collection is very limited. The average of the pairwise FST estimates between each of the 30 sample collection districts is 0.0007, with a maximum of 0.003 (Supplementary Table 1).These are extremely small differences in the European (let alone global) context. So, the British are, overall, a very homogeneous population. This is what led the researchers to use methods such as ChromoPainter/ fineStructure/ Globetrotter that can squeeze out fine-scale population structure by exploiting linkage disequilibrium. Thus, the authors are able to detect 17 main clusters of the British.
Most of the clusters are geographical, but some span different regions (e.g., the "yellow circle" cluster). The elephant in the room is the "red square" cluster which spans Central/South England. The authors write:
There is a single large cluster (red squares) that covers most of central and southern England and extends up the east coast. Notably, even at the finest level of differentiation returned by fineSTRUCTURE (53 clusters), this cluster remains largely intact and contains almost half the individuals (1,006) in our study.The authors then tried to infer the ancestry of the British clusters in terms of continental European clusters, which is to be published separately. In the plot on the right, you see the British clusters (columns) and their continental European sources (rows). The authors observe that clusters that are widely represented in Britain are likely to be older, while those that are missing in some populations are likely to be younger, because they didn't have the chance to spread across Britain. For example, a couple of Norwegian clusters are strongly represented in the Orkney islands, and these are likely to reflect Viking colonization.
The authors draw conclusions on several historical episodes of British history. The big one is the extent of Anglo-Saxon ancestry:
After the Saxon migrations, the language, place names, cereal crops and pottery styles all changed from that of the existing (Romano-British) population to those of the Saxon migrants. There has been ongoing historical and archaeological controversy about the extent to which the Saxons replaced the existing Romano-British populations. Earlier genetic analyses, based on limited samples and specific loci, gave conflicting results. With genome-wide data we can resolve this debate. Two separate analyses (ancestry profiles and GLOBETROTTER) show clear evidence in modern England of the Saxon migration, but each limits the proportion of Saxon ancestry, clearly excluding the possibility of long-term Saxon replacement. We estimate the proportion of Saxon ancestry in Cent./S England as very likely to be under 50%, and most likely in the range of 10–40%.Two other details are the lack of Danish Viking ancestry in England:
In particular, we see no clear genetic evidence of the Danish Viking occupation and control of a large part of England, either in separate UK clusters in that region, or in estimated ancestry profiles, suggesting a relatively limited input of DNA from the Danish Vikings and subsequent mixing with nearby regions, and clear evidence for only a minority Norse contribution (about 25%) to the current Orkney population.And, the absence of a unified pre-Saxon "Celtic" population. What seems to unify "Celts" is lower levels/absence of the Saxon influence, rather than belonging to a homogeneous "Celtic" population:
We saw no evidence of a general ‘Celtic’ population in non-Saxon parts of the UK. Instead there were many distinct genetic clusters in these regions, some amongst the most different in our study, in the sense of being most separated in the hierarchical clustering tree in Fig. 1. Further, the ancestry profile of Cornwall (perhaps expected to resemble other Celtic clusters) is quite different from that of the Welsh clusters, and much closer to that of Devon, and Cent./S England. However, the data do suggest that the Welsh clusters represent populations that are more similar to the early post-Ice-Age settlers of Britain than those from elsewhere in the UK.Unfortunately, the authors have decided not to make their data publicly available. This is very unfortunate, and will keep this research out of the hands of many people who would be interested in it and who would be interested in analyzing this data. I can already guess the disappointment of people of British ancestry from around the world who have a genealogical interest in tracing their British ancestors to particular areas of the UK. Apparently, the data is deposited in the EGA archive, access requires red tape, and is apparently limited to institutional researchers. Thus, this data, perhaps the richest genetic survey of any country to date, will not be fully utilized to further science.
Nature 519, 309–314 (19 March 2015) doi:10.1038/nature14230
The fine-scale genetic structure of the British population
Stephen Leslie et al.
Fine-scale genetic variation between human populations is interesting as a signature of historical demographic events and because of its potential for confounding disease studies. We use haplotype-based statistical methods to analyse genome-wide single nucleotide polymorphism (SNP) data from a carefully chosen geographically diverse sample of 2,039 individuals from the United Kingdom. This reveals a rich and detailed pattern of genetic differentiation with remarkable concordance between genetic clusters and geography. The regional genetic differentiation and differing patterns of shared ancestry with 6,209 individuals from across Europe carry clear signals of historical demographic events. We estimate the genetic contribution to southeastern England from Anglo-Saxon migrations to be under half, and identify the regions not carrying genetic material from these migrations. We suggest significant pre-Roman but post-Mesolithic movement into southeastern England from continental Europe, and show that in non-Saxon parts of the United Kingdom, there exist genetically differentiated subgroups rather than a general ‘Celtic’ population.
Link
March 15, 2015
Natural selection and ancient European DNA
A new preprint on the bioRxiv studies the same data as the recent Haak et al. paper, but focuses on natural selection in Europe. Until recently, selection could only be studied by looking at modern populations, but since selection is genetic change over time effected by the environment, it's possible that studies like this will be the norm in the future.
The new study seems to confirm the results of Wilde et al. on steppe groups, as the Yamnaya had a very low frequency of the HERC2 derived "blue eye" allele and a lower frequency of the SLC45A2 "light skin" allele than any modern Europeans. The Yamnaya seem to have been fixed for the other SLC24A5 "light skin" allele which seems to have been at high frequency in all ancient groups save the "Western Hunter Gatherers".
It seems that light pigmentation traits had already existed in pre-Indo-European Europeans (both farmers and hunter-gatherers) and so long-standing philological attempts to correlate them with the arrival of light-pigmented Indo-Europeans from the steppe (or indeed anywhere), and to contrast them with darker pre-Indo-European inhabitants of Europe were misguided. If anything, it seems that the "fairest of them all" were the Scandinavian hunter-gatherers, and a combination of light/dark pigmentation was also present in Neolithic farmers and Western Hunter Gatherers in various combinations.
It also seems that both the theory that lactose tolerance started with LBK farmers and the theory that it came to Europe from milk-drinking steppe Indo-Europeans were wrong, as this trait seems to be altogether absent in European hunter-gatherers, farmers, and Yamnaya, and make a very timid appearance in the Late neolithic/Bronze Age before shooting up in frequency to the present.
Another new development is the ability to predict "genetic height" from ancient DNA. I think this may be a little bit superfluous as you can predict "actual height" by measuring long bone lengths. On the other hand, actualized height depends not only on genetics but also on diet, disease, etc., so it's useful to look at genetic changes in such polygenic traits directly.
A big surprise was the presence of the derived EDAR allele in Swedish hunter-gatherers. This allele is very rare in modern Europeans and seems to have pleiotropic effects in East Asians. This raises the question why this allele (that was so successful in East Asians), never "took hold" in Europeans. One possibility is that it never provided an advantage to Europeans (I don't think anyone really knows what it's actually good for). Another is that Swedish hunter-gatherers simply didn't contribute much ancestry to modern Europeans and so the allele never got the chance to rise in frequency by much.
bioRxiv http://dx.doi.org/10.1101/016477
Eight thousand years of natural selection in Europe
Iain Mathieson et al.
The arrival of farming in Europe beginning around 8,500 years ago required adaptation to new environments, pathogens, diets, and social organizations. While evidence of natural selection can be revealed by studying patterns of genetic variation in present-day people, these pattern are only indirect echoes of past events, and provide little information about where and when selection occurred. Ancient DNA makes it possible to examine populations as they were before, during and after adaptation events, and thus to reveal the tempo and mode of selection. Here we report the first genome-wide scan for selection using ancient DNA, based on 83 human samples from Holocene Europe analyzed at over 300,000 positions. We find five genome-wide signals of selection, at loci associated with diet and pigmentation. Surprisingly in light of suggestions of selection on immune traits associated with the advent of agriculture and denser living conditions, we find no strong sweeps associated with immunological phenotypes. We also report a scan for selection for complex traits, and find two signals of selection on height: for short stature in Iberia after the arrival of agriculture, and for tall stature on the Pontic-Caspian steppe earlier than 5,000 years ago. A surprise is that in Scandinavian hunter-gatherers living around 8,000 years ago, there is a high frequency of the derived allele at the EDAR gene that is the strongest known signal of selection in East Asians and that is thought to have arisen in East Asia. These results document the power of ancient DNA to reveal features of past adaptation that could not be understood from analyses of present-day people.
Link (pdf)
The new study seems to confirm the results of Wilde et al. on steppe groups, as the Yamnaya had a very low frequency of the HERC2 derived "blue eye" allele and a lower frequency of the SLC45A2 "light skin" allele than any modern Europeans. The Yamnaya seem to have been fixed for the other SLC24A5 "light skin" allele which seems to have been at high frequency in all ancient groups save the "Western Hunter Gatherers".
It seems that light pigmentation traits had already existed in pre-Indo-European Europeans (both farmers and hunter-gatherers) and so long-standing philological attempts to correlate them with the arrival of light-pigmented Indo-Europeans from the steppe (or indeed anywhere), and to contrast them with darker pre-Indo-European inhabitants of Europe were misguided. If anything, it seems that the "fairest of them all" were the Scandinavian hunter-gatherers, and a combination of light/dark pigmentation was also present in Neolithic farmers and Western Hunter Gatherers in various combinations.
It also seems that both the theory that lactose tolerance started with LBK farmers and the theory that it came to Europe from milk-drinking steppe Indo-Europeans were wrong, as this trait seems to be altogether absent in European hunter-gatherers, farmers, and Yamnaya, and make a very timid appearance in the Late neolithic/Bronze Age before shooting up in frequency to the present.
Another new development is the ability to predict "genetic height" from ancient DNA. I think this may be a little bit superfluous as you can predict "actual height" by measuring long bone lengths. On the other hand, actualized height depends not only on genetics but also on diet, disease, etc., so it's useful to look at genetic changes in such polygenic traits directly.
A big surprise was the presence of the derived EDAR allele in Swedish hunter-gatherers. This allele is very rare in modern Europeans and seems to have pleiotropic effects in East Asians. This raises the question why this allele (that was so successful in East Asians), never "took hold" in Europeans. One possibility is that it never provided an advantage to Europeans (I don't think anyone really knows what it's actually good for). Another is that Swedish hunter-gatherers simply didn't contribute much ancestry to modern Europeans and so the allele never got the chance to rise in frequency by much.
bioRxiv http://dx.doi.org/10.1101/016477
Eight thousand years of natural selection in Europe
Iain Mathieson et al.
The arrival of farming in Europe beginning around 8,500 years ago required adaptation to new environments, pathogens, diets, and social organizations. While evidence of natural selection can be revealed by studying patterns of genetic variation in present-day people, these pattern are only indirect echoes of past events, and provide little information about where and when selection occurred. Ancient DNA makes it possible to examine populations as they were before, during and after adaptation events, and thus to reveal the tempo and mode of selection. Here we report the first genome-wide scan for selection using ancient DNA, based on 83 human samples from Holocene Europe analyzed at over 300,000 positions. We find five genome-wide signals of selection, at loci associated with diet and pigmentation. Surprisingly in light of suggestions of selection on immune traits associated with the advent of agriculture and denser living conditions, we find no strong sweeps associated with immunological phenotypes. We also report a scan for selection for complex traits, and find two signals of selection on height: for short stature in Iberia after the arrival of agriculture, and for tall stature on the Pontic-Caspian steppe earlier than 5,000 years ago. A surprise is that in Scandinavian hunter-gatherers living around 8,000 years ago, there is a high frequency of the derived allele at the EDAR gene that is the strongest known signal of selection in East Asians and that is thought to have arisen in East Asia. These results document the power of ancient DNA to reveal features of past adaptation that could not be understood from analyses of present-day people.
Link (pdf)
March 14, 2015
Bottleneck in human Y-chromosomes in the last 10,000 years.
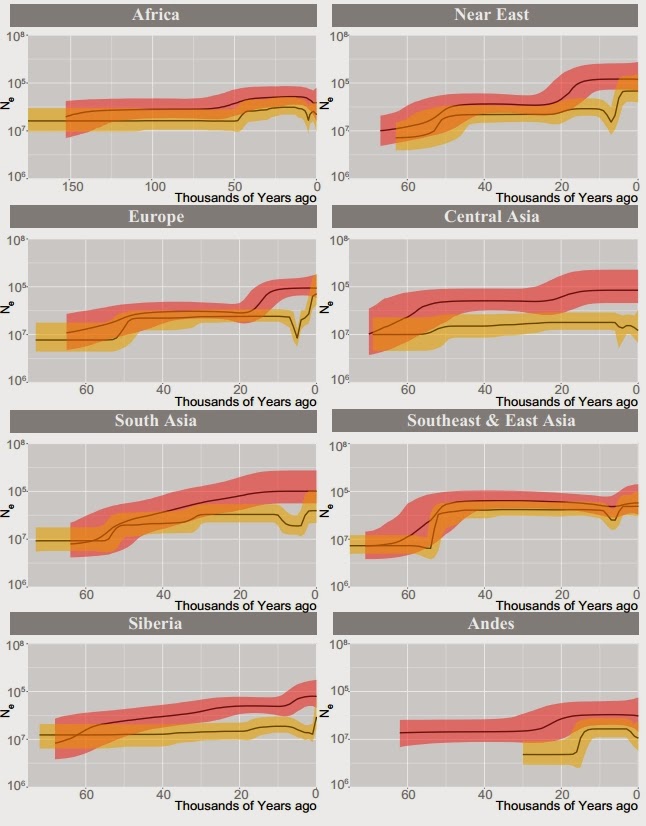
A very exciting new paper has just been published in Genome Research on 456 full sequence Y-chromosomes from around the world. The authors date the MRCA of Y-chromosomes ("Y chromosome Adam") to 254 (95% CI 192–307) kya, find coalescences of major non-African haplogroups to 47–52 kya (which clearly corresponds to the Upper Paleolithic revolution), but also infer a second bottleneck that occurred in the last 10 thousand years.
The contrast (left) between mtDNA (red) and Y-chromosome (yellow) coalescences is quite noticeable. The little "dip" in the yellow curve in many regions on the right of the various regional plots corresponds to a the second bottleneck event (that was really not "one" event, but rather shows that many modern men descend from a small number of "patriarchs" of the Neolithic and Bronze Age worlds. The "when" of the dip is important:

Most human mythologies contain stories of "first men" and eponymous founders of nations; these were often ridiculed in recent times as invented stories whose purpose was to engender social cohesion through a story of shared descent. But, now it seems that these stories were at least in part true, and such ultra-prolific patriarchs do indeed stand at the beginning of many later lines of descent.
Figure 1 from the paper is an extremely useful overview of human Y-chromosome phylogeny.
The split between DT and B2'5 is placed at around ~100 thousand years ago. This corresponds perfectly (in my opinion) to the Out-of-Africa event from which most Eurasian men are probably descended. For the next thirty thousand years, Eurasians were probably confined to Arabia and the Middle East. The next major event is the foundation of the unambiguously Eurasian CT lineage ~70 thousand years ago (coinciding with the Toba eruption and the onset of super arid conditions at the onset of MIS 4). And, the final event of the "grand picture" of Eurasian prehistory was the Upper Paleolithic at ~50 thousand years ago, when Eurasians finally got the "tech" to exhibit complex behavior, invent new tools, conquer diverse environments and ultimately colonize the entire planet while driving Eurasian archaics to extinction.
An important detail in this grand picture is the fact that the authors estimate the coalescence date between D and E1'4 to ~70,000 years, coinciding with the C/GT split from the same time. These lineages are all found in Eurasia, but only E1'4 is found in Africa. I think this points clearly to back-migration of Eurasians into Africa, perhaps as environmental refugees following the c. 70kya Arabian ecological catastrophe. In any case, the fact that two separate Eurasian-specific lineages (CT and D) coalesce to ~70kya destroys the theory that the spread of modern humans into Eurasia happened together with UP-related technologies, a theory that was already on its last legs given the evidence that pre-UP admixture with Neandertals had taken place (as such admixture would have been impossible if pre-UP Eurasians were not already present outside of Africa at that time).
Genome Research doi:10.1101/gr.186684.114
A recent bottleneck of Y chromosome diversity coincides with a global change in culture
Monika Karmin et al.
It is commonly thought that human genetic diversity in non-African populations was shaped primarily by an out-of-Africa dispersal 50–100 thousand yr ago (kya). Here, we present a study of 456 geographically diverse high-coverage Y chromosome sequences, including 299 newly reported samples. Applying ancient DNA calibration, we date the Y-chromosomal most recent common ancestor (MRCA) in Africa at 254 (95% CI 192–307) kya and detect a cluster of major non-African founder haplogroups in a narrow time interval at 47–52 kya, consistent with a rapid initial colonization model of Eurasia and Oceania after the out-of-Africa bottleneck. In contrast to demographic reconstructions based on mtDNA, we infer a second strong bottleneck in Y-chromosome lineages dating to the last 10 ky. We hypothesize that this bottleneck is caused by cultural changes affecting variance of reproductive success among males.
Link
The contrast (left) between mtDNA (red) and Y-chromosome (yellow) coalescences is quite noticeable. The little "dip" in the yellow curve in many regions on the right of the various regional plots corresponds to a the second bottleneck event (that was really not "one" event, but rather shows that many modern men descend from a small number of "patriarchs" of the Neolithic and Bronze Age worlds. The "when" of the dip is important:

Most human mythologies contain stories of "first men" and eponymous founders of nations; these were often ridiculed in recent times as invented stories whose purpose was to engender social cohesion through a story of shared descent. But, now it seems that these stories were at least in part true, and such ultra-prolific patriarchs do indeed stand at the beginning of many later lines of descent.
The split between DT and B2'5 is placed at around ~100 thousand years ago. This corresponds perfectly (in my opinion) to the Out-of-Africa event from which most Eurasian men are probably descended. For the next thirty thousand years, Eurasians were probably confined to Arabia and the Middle East. The next major event is the foundation of the unambiguously Eurasian CT lineage ~70 thousand years ago (coinciding with the Toba eruption and the onset of super arid conditions at the onset of MIS 4). And, the final event of the "grand picture" of Eurasian prehistory was the Upper Paleolithic at ~50 thousand years ago, when Eurasians finally got the "tech" to exhibit complex behavior, invent new tools, conquer diverse environments and ultimately colonize the entire planet while driving Eurasian archaics to extinction.
An important detail in this grand picture is the fact that the authors estimate the coalescence date between D and E1'4 to ~70,000 years, coinciding with the C/GT split from the same time. These lineages are all found in Eurasia, but only E1'4 is found in Africa. I think this points clearly to back-migration of Eurasians into Africa, perhaps as environmental refugees following the c. 70kya Arabian ecological catastrophe. In any case, the fact that two separate Eurasian-specific lineages (CT and D) coalesce to ~70kya destroys the theory that the spread of modern humans into Eurasia happened together with UP-related technologies, a theory that was already on its last legs given the evidence that pre-UP admixture with Neandertals had taken place (as such admixture would have been impossible if pre-UP Eurasians were not already present outside of Africa at that time).
Genome Research doi:10.1101/gr.186684.114
A recent bottleneck of Y chromosome diversity coincides with a global change in culture
Monika Karmin et al.
It is commonly thought that human genetic diversity in non-African populations was shaped primarily by an out-of-Africa dispersal 50–100 thousand yr ago (kya). Here, we present a study of 456 geographically diverse high-coverage Y chromosome sequences, including 299 newly reported samples. Applying ancient DNA calibration, we date the Y-chromosomal most recent common ancestor (MRCA) in Africa at 254 (95% CI 192–307) kya and detect a cluster of major non-African founder haplogroups in a narrow time interval at 47–52 kya, consistent with a rapid initial colonization model of Eurasia and Oceania after the out-of-Africa bottleneck. In contrast to demographic reconstructions based on mtDNA, we infer a second strong bottleneck in Y-chromosome lineages dating to the last 10 ky. We hypothesize that this bottleneck is caused by cultural changes affecting variance of reproductive success among males.
Link
March 11, 2015
Genetic pacification of Western Europeans (?)
Evolutionary Psychology
www.epjournal.net – 2015. 13(1): 230-243
Western Europe, State Formation, and Genetic Pacification
Peter Frost, Henry C. Harpending
Through its monopoly on violence, the State tends to pacify social relations. Such pacification proceeded slowly in Western Europe between the 5th and 11th centuries, being hindered by the rudimentary nature of law enforcement, the belief in a man’s right to settle personal disputes as he saw fit, and the Church’s opposition to the death penalty. These hindrances began to dissolve in the 11th century with a consensus by Church and State that the wicked should be punished so that the good may live in peace. Courts imposed the death penalty more and more often and, by the late Middle Ages, were condemning to death between 0.5 and 1.0% of all men of each generation, with perhaps just as many offenders dying at the scene of the crime or in prison while awaiting trial. Meanwhile, the homicide rate plummeted from the 14th century to the 20th. The pool of violent men dried up until most murders occurred under conditions of jealousy, intoxication, or extreme stress. The decline in personal violence is usually attributed to harsher punishment and the longer-term effects of cultural conditioning. It may also be, however, that this new cultural environment selected against propensities for violence.
Link (pdf)
Western Europe, State Formation, and Genetic Pacification
Peter Frost, Henry C. Harpending
Through its monopoly on violence, the State tends to pacify social relations. Such pacification proceeded slowly in Western Europe between the 5th and 11th centuries, being hindered by the rudimentary nature of law enforcement, the belief in a man’s right to settle personal disputes as he saw fit, and the Church’s opposition to the death penalty. These hindrances began to dissolve in the 11th century with a consensus by Church and State that the wicked should be punished so that the good may live in peace. Courts imposed the death penalty more and more often and, by the late Middle Ages, were condemning to death between 0.5 and 1.0% of all men of each generation, with perhaps just as many offenders dying at the scene of the crime or in prison while awaiting trial. Meanwhile, the homicide rate plummeted from the 14th century to the 20th. The pool of violent men dried up until most murders occurred under conditions of jealousy, intoxication, or extreme stress. The decline in personal violence is usually attributed to harsher punishment and the longer-term effects of cultural conditioning. It may also be, however, that this new cultural environment selected against propensities for violence.
Link (pdf)
March 10, 2015
DNA of 17th century African slaves frome the Caribbean
PNAS doi: 10.1073/pnas.1421784112
Genome-wide ancestry of 17th-century enslaved Africans from the Caribbean
Hannes Schroeder, María C. Ávila-Arcos et al.
Between 1500 and 1850, more than 12 million enslaved Africans were transported to the New World. The vast majority were shipped from West and West-Central Africa, but their precise origins are largely unknown. We used genome-wide ancient DNA analyses to investigate the genetic origins of three enslaved Africans whose remains were recovered on the Caribbean island of Saint Martin. We trace their origins to distinct subcontinental source populations within Africa, including Bantu-speaking groups from northern Cameroon and non-Bantu speakers living in present-day Nigeria and Ghana. To our knowledge, these findings provide the first direct evidence for the ethnic origins of enslaved Africans, at a time for which historical records are scarce, and demonstrate that genomic data provide another type of record that can shed new light on long-standing historical questions.
Link
Genome-wide ancestry of 17th-century enslaved Africans from the Caribbean
Hannes Schroeder, María C. Ávila-Arcos et al.
Between 1500 and 1850, more than 12 million enslaved Africans were transported to the New World. The vast majority were shipped from West and West-Central Africa, but their precise origins are largely unknown. We used genome-wide ancient DNA analyses to investigate the genetic origins of three enslaved Africans whose remains were recovered on the Caribbean island of Saint Martin. We trace their origins to distinct subcontinental source populations within Africa, including Bantu-speaking groups from northern Cameroon and non-Bantu speakers living in present-day Nigeria and Ghana. To our knowledge, these findings provide the first direct evidence for the ethnic origins of enslaved Africans, at a time for which historical records are scarce, and demonstrate that genomic data provide another type of record that can shed new light on long-standing historical questions.
Link
March 09, 2015
Sveti Ivan relics Middle Eastern, mid-1st century AD
Irrespective of one's religious beliefs, a genome from the mid-1st century AD would be interesting. I am personally in favor of the scientific study of saints' relics.
Bulgarian bones could be John the Baptist's, scientists say
Bulgarian bones could be John the Baptist's, scientists say
So when Bulgarian archeologists announced in 2010 that they had found the bones of John the Baptist, Tom Higham was skeptical.St. John's father Zechariah belonged to to the Aaronic line of priests. While modern Jewish priests (Cohanim) belong to multiple Y-chromosomal lineages, I think it's a good bet that at least one or a few of these lineages could be traced to Jewish priests of 2,000 years ago (leaving the question of the ultimate Aaronic lineage open). If the remains from Sveti Ivan island match one of these lineages, then this would be a powerful piece of evidence in favor of their attribution to St. John the Baptist.
He got a surprise.
Higham, an Oxford University scientist and an atheist who doesn't believe in "any kind of religion or God or anything like that," was asked to test six small bone fragments found on an island named Sveti Ivan - St. John.
The bones turned out to be from a man who lived in the Middle East at the same time as Jesus, Higham said.
"We got a date that was exactly where it should be, right in the middle of the first century," said Higham, a radiocarbon dating expert.
...
DNA testing by colleagues at the University of Copenhagen suggested that the person was most likely to have been from the Middle East, he said.
"We have a complete genome. It's possible that we could step this a step further and see if there is any similarity," in the genetic material of all the relics.
March 06, 2015
Craniofacial plasticity in ancient Peru
Anthropologischer Anzeiger doi:10.1127/anthranz/2015/0458
Craniofacial plasticity in ancient Peru
Jessica H. Stone; Kristen Chew; Ann H. Ross; John W. Verano
Numerous studies have utilized craniometric data to explore the roles of genetic diversity and environment in human cranial shape variation. Peru is a particularly interesting region to examine cranial variation due to the wide variety of high and low altitude ecological zones, which in combination with rugged terrain have created isolated populations with vastly different physiological adaptations. This study examines seven samples from throughout Peru in an effort to understand the contributions of environmental adaptation and genetic relatedness to craniofacial variation at a regional scale. Morphological variation was investigated using a canonical discriminant analysis and Mahalanobis D2 analysis. Results indicate that all groups are significantly different from one another with the closest relationship between Yauyos and Jahuay, two sites that are located geographically close in central Peru but in very different ecozones. The relationship between latitude/longitude and face shape was also examined with a spatial autocorrelation analysis (Moran’s I) using ArcMap and show that there is significant spatial patterning for facial measures and geographic location suggesting that there is an association between biological variation and geographic location.
Link
Craniofacial plasticity in ancient Peru
Jessica H. Stone; Kristen Chew; Ann H. Ross; John W. Verano
Numerous studies have utilized craniometric data to explore the roles of genetic diversity and environment in human cranial shape variation. Peru is a particularly interesting region to examine cranial variation due to the wide variety of high and low altitude ecological zones, which in combination with rugged terrain have created isolated populations with vastly different physiological adaptations. This study examines seven samples from throughout Peru in an effort to understand the contributions of environmental adaptation and genetic relatedness to craniofacial variation at a regional scale. Morphological variation was investigated using a canonical discriminant analysis and Mahalanobis D2 analysis. Results indicate that all groups are significantly different from one another with the closest relationship between Yauyos and Jahuay, two sites that are located geographically close in central Peru but in very different ecozones. The relationship between latitude/longitude and face shape was also examined with a spatial autocorrelation analysis (Moran’s I) using ArcMap and show that there is significant spatial patterning for facial measures and geographic location suggesting that there is an association between biological variation and geographic location.
Link
March 03, 2015
Oxhide ingots in Scandinavian rock art
Antiquity / Volume 89 / Issue 343 / February 2015, pp 191-209
Representations of oxhide ingots in Scandinavian rock art: the sketchbook of a Bronze Age traveller?
Johan Ling and Zofia Stos-Gale
Bronze Age trade networks across Europe and the Mediterranean are well documented; Baltic amber and bronze metalwork were particularly valued commodities. Here it is argued that demand for copper and tin led to changes in Scandinavian trade routes around 1600 BC, which can be linked to the appearance of figurative rock art images in southern Scandinavia. Images identified as oxhide ingots have been discovered in Sweden and suggest that people from Scandinavia were familiar with this characteristically Mediterranean trading commodity. Using trace element and lead isotope analysis, the authors argue that some bronze tools excavated in Sweden could have been made of Cypriot copper; these two discoveries suggest that Scandinavians were travelling to the Mediterranean, rather than acting through a middle man.
Link
Representations of oxhide ingots in Scandinavian rock art: the sketchbook of a Bronze Age traveller?
Johan Ling and Zofia Stos-Gale
Bronze Age trade networks across Europe and the Mediterranean are well documented; Baltic amber and bronze metalwork were particularly valued commodities. Here it is argued that demand for copper and tin led to changes in Scandinavian trade routes around 1600 BC, which can be linked to the appearance of figurative rock art images in southern Scandinavia. Images identified as oxhide ingots have been discovered in Sweden and suggest that people from Scandinavia were familiar with this characteristically Mediterranean trading commodity. Using trace element and lead isotope analysis, the authors argue that some bronze tools excavated in Sweden could have been made of Cypriot copper; these two discoveries suggest that Scandinavians were travelling to the Mediterranean, rather than acting through a middle man.
Link
March 02, 2015
mtDNA from Lengyel culture in Poland
PLoS ONE 10(2): e0118316. doi:10.1371/journal.pone.0118316
Between the Baltic and Danubian Worlds: The Genetic Affinities of a Middle Neolithic Population from Central Poland
Wiesław Lorkiewicz et al.
For a long time, anthropological and genetic research on the Neolithic revolution in Europe was mainly concentrated on the mechanism of agricultural dispersal over different parts of the continent. Recently, attention has shifted towards population processes that occurred after the arrival of the first farmers, transforming the genetically very distinctive early Neolithic Linear Pottery Culture (LBK) and Mesolithic forager populations into present-day Central Europeans. The latest studies indicate that significant changes in this respect took place within the post-Linear Pottery cultures of the Early and Middle Neolithic which were a bridge between the allochthonous LBK and the first indigenous Neolithic culture of north-central Europe—the Funnel Beaker culture (TRB). The paper presents data on mtDNA haplotypes of a Middle Neolithic population dated to 4700/4600–4100/4000 BC belonging to the Brześć Kujawski Group of the Lengyel culture (BKG) from the Kuyavia region in north-central Poland. BKG communities constituted the border of the “Danubian World” in this part of Europe for approx. seven centuries, neighboring foragers of the North European Plain and the southern Baltic basin. MtDNA haplogroups were determined in 11 individuals, and four mtDNA macrohaplogroups were found (H, U5, T, and HV0). The overall haplogroup pattern did not deviate from other post-Linear Pottery populations from central Europe, although a complete lack of N1a and the presence of U5a are noteworthy. Of greatest importance is the observed link between the BKG and the TRB horizon, confirmed by an independent analysis of the craniometric variation of Mesolithic and Neolithic populations inhabiting central Europe. Estimated phylogenetic pattern suggests significant contribution of the post-Linear BKG communities to the origin of the subsequent Middle Neolithic cultures, such as the TRB.
Link
Between the Baltic and Danubian Worlds: The Genetic Affinities of a Middle Neolithic Population from Central Poland
Wiesław Lorkiewicz et al.
For a long time, anthropological and genetic research on the Neolithic revolution in Europe was mainly concentrated on the mechanism of agricultural dispersal over different parts of the continent. Recently, attention has shifted towards population processes that occurred after the arrival of the first farmers, transforming the genetically very distinctive early Neolithic Linear Pottery Culture (LBK) and Mesolithic forager populations into present-day Central Europeans. The latest studies indicate that significant changes in this respect took place within the post-Linear Pottery cultures of the Early and Middle Neolithic which were a bridge between the allochthonous LBK and the first indigenous Neolithic culture of north-central Europe—the Funnel Beaker culture (TRB). The paper presents data on mtDNA haplotypes of a Middle Neolithic population dated to 4700/4600–4100/4000 BC belonging to the Brześć Kujawski Group of the Lengyel culture (BKG) from the Kuyavia region in north-central Poland. BKG communities constituted the border of the “Danubian World” in this part of Europe for approx. seven centuries, neighboring foragers of the North European Plain and the southern Baltic basin. MtDNA haplogroups were determined in 11 individuals, and four mtDNA macrohaplogroups were found (H, U5, T, and HV0). The overall haplogroup pattern did not deviate from other post-Linear Pottery populations from central Europe, although a complete lack of N1a and the presence of U5a are noteworthy. Of greatest importance is the observed link between the BKG and the TRB horizon, confirmed by an independent analysis of the craniometric variation of Mesolithic and Neolithic populations inhabiting central Europe. Estimated phylogenetic pattern suggests significant contribution of the post-Linear BKG communities to the origin of the subsequent Middle Neolithic cultures, such as the TRB.
Link
Y chromosome mutation rate: 0.82x10-9
This is in Russian, but seems to be using Anzick-1, Ust-Ishim, K14 to get the mutation rate. Fu et al. who just used Ust Ishim got 0.7-0.9 for this which seems very similar, and also identical to the 0.82x10-9 value of Poznik et al. So, for the time being this seems like the value to use, although tighter confidence intervals would be welcome.
The Russian Journal of Genetic Genealogy (Русская версия), Vol 6, No 2 (2014)/Vol 7, No 1 (2015)
Константа скорости SNP мутаций Y-хромосомы по данным полного секвенирования
Дмитрий Адамов, Владимир Гурьянов, Сергей Каржавин, Владимир Таганкин, Вадим Урасин
Abstract
Накопление данных тестирования BigY, FGC, с одной стороны, публикация сиквенсов Y-хромосомы древних образцов Anzick-1, Ust-Ishim, K14, с другой, дает возможность оценить среднюю скорость однонуклеотидных (SNP) мутаций. Авторы разработали собственный метод отбора истинных мутаций в современных и древних образцах и несколькими способами определили с высокой точностью константу скорости SNP мутаций - 0.82·10-9 в год на п.н.о. (95% CI: (0.70-0.94) ·10-9).
Link
The Russian Journal of Genetic Genealogy (Русская версия), Vol 6, No 2 (2014)/Vol 7, No 1 (2015)
Константа скорости SNP мутаций Y-хромосомы по данным полного секвенирования
Дмитрий Адамов, Владимир Гурьянов, Сергей Каржавин, Владимир Таганкин, Вадим Урасин
Abstract
Накопление данных тестирования BigY, FGC, с одной стороны, публикация сиквенсов Y-хромосомы древних образцов Anzick-1, Ust-Ishim, K14, с другой, дает возможность оценить среднюю скорость однонуклеотидных (SNP) мутаций. Авторы разработали собственный метод отбора истинных мутаций в современных и древних образцах и несколькими способами определили с высокой точностью константу скорости SNP мутаций - 0.82·10-9 в год на п.н.о. (95% CI: (0.70-0.94) ·10-9).
Link
March 01, 2015
8,000 year old wheat in Britain
Britain received farming later than most of Europe, but perhaps it received one of the products of farming well before any farmers set foot on the island. I've always wondered if news (and at least some products) of the agricultural revolution spread far and wide before the revolution itself did. Did foragers at the northwestern end of Europe hear stories of the strange new people that had already appeared 8,000 years ago on the opposite end of the continent?
Link
Was this an isolated incident or will we be finding wheat elsewhere in pre-farming Europe?
Science 27 February 2015: Vol. 347 no. 6225 pp. 998-1001
Sedimentary DNA from a submerged site reveals wheat in the British Isles 8000 years ago
Oliver Smith et al.
The Mesolithic-to-Neolithic transition marked the time when a hunter-gatherer economy gave way to agriculture, coinciding with rising sea levels. Bouldnor Cliff, is a submarine archaeological site off the Isle of Wight in the United Kingdom that has a well-preserved Mesolithic paleosol dated to 8000 years before the present. We analyzed a core obtained from sealed sediments, combining evidence from microgeomorphology and microfossils with sedimentary ancient DNA (sedaDNA) analyses to reconstruct floral and faunal changes during the occupation of this site, before it was submerged. In agreement with palynological analyses, the sedaDNA sequences suggest a mixed habitat of oak forest and herbaceous plants. However, they also provide evidence of wheat 2000 years earlier than mainland Britain and 400 years earlier than proximate European sites. These results suggest that sophisticated social networks linked the Neolithic front in southern Europe to the Mesolithic peoples of northern Europe.Link
Two observations on the ancestry of Armenians
 I was thinking a bit on how to interpret the findings of the new Haber et al. preprint, and especially the idea that "29% of the Armenian ancestry may originate from an ancestral population best represented by Neolithic Europeans." I looked at the globe13 proportions, and strangely enough, I had estimated that the three Armenian samples (Armenian_D, Armenians, and Armenians_15_Y) have 28-29% of the Mediterranean component that is modal in Sardinians. This seems like a curious coincidence which has raised my confidence that Haber et al. is picking something real.
I was thinking a bit on how to interpret the findings of the new Haber et al. preprint, and especially the idea that "29% of the Armenian ancestry may originate from an ancestral population best represented by Neolithic Europeans." I looked at the globe13 proportions, and strangely enough, I had estimated that the three Armenian samples (Armenian_D, Armenians, and Armenians_15_Y) have 28-29% of the Mediterranean component that is modal in Sardinians. This seems like a curious coincidence which has raised my confidence that Haber et al. is picking something real.
Looking back at my inferences of Armenian ancestry, it seems (according to globe13) to come completely from West_Asian, Mediterranean, and Southwest_Asian. The Mediterranean component seems real enough as it seems to match Sardinians/early European farmers well. I am not so sure about the Southwest Asian component which is modal in Yemen Jews and may represent population-specific drift in relatively recent Arabians. The West_Asian component is bimodal in Caucasus and Gedrosia, so it can't be the result of a very drifted population in either region (unless there is spooky action at a distance).
Another curious finding is the lack of North_European in a latitudinal "column" of populations from the Yemen, through the Levant to the South Caucasus (Georgians and Armenians). It seems that North_European is the only one of the four major Caucasoid components that Armenians lack to any important degree. There is a rather abrupt change between the South Caucasus (~1%) and the North Caucasus (15-20%). It seems that the Greater Caucasus did act like a barrier to gene flow. The K=4 analysis of the same dataset that produced K=13 (globe13) also shows the same barrier: all three Armenian samples and Georgians have ~0% of "Amerindian" (which is surely correlated to "Ancient North Eurasian" ancestry and via it with North_European), but North Caucasians and Europeans have 4-10%. It's clear that this influence did not cross the Greater Caucasus, as Armenians and Georgians lack it.
Subscribe to:
Comments (Atom)