An interesting problem in population genetics is the following: how often are two individuals from a population A more similar to each other than either of them is to an individual from another population B?
Suppose you have a similarity function for individuals, sim(a, b)
(In my experiments I will use identity-by-state (IBS) as calculated by PLINK (--cluster --matrix) as a measure of similarity, but any symmetrical similarity function (that is, sim(a,b)=sim(b,a)) will do.)
We want to calculate the rate at which the following condition occurs:
If this condition holds, then a and a' from population A are more similar to each other than either of them is to an individual b from population B. We then say that the trio of individuals is concordant.
I will use the indicator function I(a, a', b) = 1 in case of a concordant trio, and =0 otherwise.
I can then estimate the probability of concordance, if I have n individuals from A and m from B as follows:
The rationale behind this formula is straightforward: we are counting the number of all concordant trios, and dividing by n(n-1)m/2 since there are n(n-1)/2 pairs of individuals from A, and each pair is compared against all m individuals from B.
The expected value of this concordance ratio can vary between 0.25 and 1:
- It is 1 if the two populations are so well-differentiated so that every trio is concordant.
- On the other hand, if the two populations are genetically identical, then each similarity comparison is equivalent to a coin toss (probability = 0.5) and we are testing this condition for two different individuals from A: hence the probability of concordance for each trio is 0.5*0.5 = 0.25.
In a finite sample of individuals it is possible that the concordance ratio estimate may actually be lower than 0.25.
An interesting property of the concordance ratio is its asymmetry, that is:
We will see how this property gives some useful insight in some of the following examples.
#1. Han vs. Yoruba
This is based on the Stanford HGDP set, including only individuals as recommended by Rosenberg (2006) in his H952 set. For each experiment only SNPs with at least a 99% genotyping rate have been retained.
The first experiment is designed to showcase the concordance ratio in two well-differentiated human populations, 44 Han Chinese and 21 Yoruba Nigerians. The analysis is based on 617,602 SNPs.
That is, two Han Chinese are always more similar to each other than to a Yoruba and vice versa.
I repeated the experiment, sequentially thinning the market set randomly by a factor of 10 using PLINK's --thin 0.1 argument. Concordance remained 1 for ~61k, ~6k, and ~600 markers, and became different than 1 (but still greater than 0.95) with 50 SNPs.
Genome-wide, or, across a sizeable number of markers concordance of Han vs. Yoruba and vice versa is perfect, while for a few random SNPs it may not be.
#2. Britons vs. Mexicans
In the next experiment, I have used 90 Britons (GBR) and 69 Mexican Americans from Los Angeles (MXL) from the 1000 Genomes Project. I have included only SNPs with 99%+ genotyping rate that are also included in the HGDP Stanford data, for a total of 351,521 markers.
Notice the previously mentioned asymmetry: two Britons are virtually always closer to each other than they are to a Mexican, but a Mexican is sometimes closer to a Briton than he is to a fellow Mexican. This is due to the fact that Mexicans have variable European admixture, so a substantially European-admixed Mexican may be closer to a Briton than he is to one of his substantially Amerindian-admixed compatriots.
#3: Various Europeans
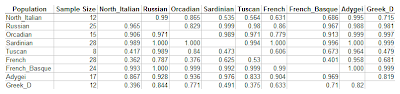
In the final experiment, I included HGDP European populations, together with 12 Dodecad Project Greeks. The analysis is based on 492,176 SNPs. Each row in the following table represents the first argument of the concordance ratio function, and each column the second one.
Here is a way to read the table, using Greek_D as an example:
- The last row represents a test in which pairs of Greeks were compared against individuals from the population of each column. These comparisons were concordant 84.4% of the time against Russians (the most distant population), and 37.5% of the time against Tuscans (the closest one).
- The last column represents in which Greeks were used as an "outgroup" for comparison against pairs of individuals from each row. These comparisons were concordant (~1) for most populations, except the Tuscan (47.9%), North Italian (71.5%), and Adygei (81.9%)
- With a pair of French individuals against individuals from other populations, concordance ranged between 36.2% (for North Italians) to 95.8% (for Adygei)
- When French individuals were used as an outgroup to compare against pairs of individuals from other populations, concordance ranged between 63.1% (for North Italians) to 100% (for Sardinians). The latter means that a pair of Sardinians is always closer to each other than to a French sample (or, at least, an HGDP French one).
Conclusion
The study of concordance is an interesting thought experiment that illustrates how genome-wide comparisons between individuals show the following:
- Two individuals from a homogeneous population are virtually always more similar to each other than to an individual from a genetically differentiated population
- Two individuals from a population may, or may not, be more similar to each other than to an individual from a genetically related population
- More variable populations are usually more discordant with respect to other populations, whereas very homogeneous populations tend to be concordant
The concordance ratio is useful for personal genomics customers, as it puts their IBS similarities to various other individuals in perspective. For example, a Greek should not be surprised if he matches a particular Tuscan more than he does a fellow Greek, nor should he seek mysterious Italian ancestors because of it, as such a discordant result occurs frequently.
It is different from techniques such as PCA which allow the separability of individuals from different populations by projecting them on a number of dimensions, the first few of which are usually correlated with the inter-population fraction of genetic diversity. Hence, individuals that appear well-separated on a few PCA dimensions may in fact be overall more genetically similar to individuals from other populations across the full marker set. The concordance ratio avoids any accusation of privileging aspects of the genome (the ones that differentiate populations), as it is based on a single genome-wide similarity function for individuals.
It is also different from clustering algorithms such as ADMIXTURE that infer allele frequencies in putative ancestral populations, again implicitly using markers with high frequency differences to estimate admixture proportions. Hence, a "match" in a marker with low population differentiation is treated differently as a source of evidence than a match in a marker of strong population differentiation. The concordance ratio avoids this issue by using a single similarity function for individuals that does not privilege one marker over another.
R Code
- A symmetrical similarity matrix, as output by plink --matrix --cluster command. Any similarity matrix file in the PLINK MIBS format will do.
- A file in which each row has a population name and the number of individuals from that population.
North_Italian 12
Russian 25
Orcadian 15
Sardinian 28
Tuscan 8
French 28
French_Basque 24
Adygei 17
Greek_D 12
Of course, individuals must appear in that order in the plink file, i.e., first the 12 North Italians, then the 25 Russians, etc.
Assuming you have such a file, e.g., in binary BED/BIM/FAM format, you first calculate the IBS matrix:
plink --matrix --cluster --bfile datafile
This creates a plink.mibs file. Then, in R, after changing to the appropriate directory, where plink.mibs, count.txt and the source code gamma.r is, you enter:
source("gamma.r")
gamma(simfile="plink.mibs", popfile="count.txt")
PS: The concordance ratio should not be confused with Witherspoon's ω fraction. That is defined by comparing all pairs of between- and within- population distances, and ranges between 0 (highest concordance in my terminology) and 0.5 (lowest concordance). The concordance ratio, on the other hand, tests all possible trios of individuals, and it also has the asymmetrical property explained above.






7 comments:
Very interesting. The most useful new concept in statistics that I've learned in a long time.
Interesting.
Instead of looking at probability of individual a's state being closer to a-prime than b, I think a more elegant way is to examine frequencies of states. As you have values such as .99 between Russian and North Italian, this would suggest that the probabilities too-easily become pegged near 1.
Further, there is no theoretical reason why more diverse populations should be less differentiated than populations who are homogeneous - if anything, they should be more, as that population contains more unique diversity. You may be interested in Jost's D as a model-free statistic for measuring population subdivision. His derivation is quite convincing.
The other comment I would have is that the advantage of ADMIXTURE is that it allows the calculation of individual proportional ancestry, which I'm not sure a concordance based approach would allow. Please correct me if I'm wrong.
Instead of looking at probability of individual a's state being closer to a-prime than b, I think a more elegant way is to examine frequencies of states.
Each individual has a unique state, so you'll have to elaborate what you mean by "frequencies of states"
As you have values such as .99 between Russian and North Italian, this would suggest that the probabilities too-easily become pegged near 1.
I don't know what you mean by "two easily". Two Italians are always closer to each other than to a Russian, that's neither easy or not-easy, it's a fact. And, there are populations for which this is not the case.
Further, there is no theoretical reason why more diverse populations should be less differentiated than populations who are homogeneous - if anything, they should be more, as that population contains more unique diversity.
The theoretical reason is a simple counter-example. A population of clones (zero diversity) would always have a concordance ratio of 1 vis a vis any other population.
The other comment I would have is that the advantage of ADMIXTURE is that it allows the calculation of individual proportional ancestry, which I'm not sure a concordance based approach would allow. Please correct me if I'm wrong.
That's like saying the advantage of a plane is that it can fly, which a submarine cannot. The two techniques address completely different problems.
Each individual has a unique state, so you'll have to elaborate what you mean by "frequencies of states"
Ah. I think I've misunderstood you. I was thinking this was something applied per-site (or per-loci), and then averaged across sites. Am I incorrect?
I don't know what you mean by "two easily". Two Italians are always closer to each other than to a Russian, that's neither easy or not-easy, it's a fact. And, there are populations for which this is not the case.
If the statistic is pegged near 1 for two populations so close together in terms of genomic composition, it doesn't have much higher to go for populations that are more distantly related (e.g., Russians and Yorubians). This would make it poor for measuring differentiation.
I'm glad you agree with me re:ADMIXTURE - that was my point as well, that ADMIXTURE (and similar programs) have a very different aim.
If the statistic is pegged near 1 for two populations so close together in terms of genomic composition, it doesn't have much higher to go for populations that are more distantly related (e.g., Russians and Yorubians). This would make it poor for measuring differentiation.
It doesn't measure differentiation across the entire range of Homo sapiens; it can be seen as marking a set of populations whose members can be genomically closer to a given population, and ordering those.
Hi, in the future could you make a table with all populations, and also if possible between dodecad members ?
Between Dodecad members is not possible, because this is a between-population measure.
As for that big table, I may eventually do it, even though it's quite time consuming to merge the various files, calculate the IBS matrix, and run the gamma calculation.
Post a Comment