It seems that positive selection studies are coming out in droves; you can use the search function ("positive selection") to find a bunch of them reported on this blog.
The newest study appears as a preprint in the American Journal of Human Genetics and is about a gene called human caspase-12 (CASP12) which has two variants: the active one is the original form, but the inactive one seems to have undergone positive selection beginning 60-100ky ago.
It is suggested that the inactive form confers some advantage against sepsis, which was also implicated in a frequent mtDNA haplogroup recently.
The distribution of the two versions of the gene...
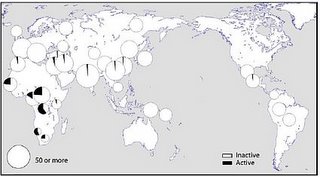
... shows that the active version is largely confined to Sub-Saharan Africa. This parallels the situation in Microcephalin and ASPM.
The highest frequencies of the active version are in the Mbuti (0.60) and the San (0.57). These populations have a high frequency of non-M168 related Y-chromosomes and non-L3 related mtDNA and are thus descended from the most ancient African populations (which I have called "Paleoafricans"), rather than the more recent emergence of an population in east Africa ("Afrasians") which spawned the Eurasians and assimilated most Paleoafricans during its spread within Africa. In this regard, it is interesting that an east African population from Kenya shows the minimum (0.04) frequency of the active gene of CASP12.
American Journal of Human Genetics (in press)
Spread of an inactive form of caspase-12 in humans due to recent
positive selection
Yali Xue et al.
Abstract
The human caspase-12 gene is polymorphic for the presence or absence of a stop codon, resulting in the occurrence of both active (ancestral) and inactive (derived) forms of the gene in the population. It has previously been shown that carriers of the inactive gene are more resistant to severe sepsis. We have now investigated whether the inactive form has spread because of neutral drift or positive selection. We determined its distribution in a worldwide sample of 52 populations and re-sequenced the gene in 77 individuals from the HapMap Yoruba, Han Chinese and European populations. There is strong evidence for positive selection from low diversity, skewed allele frequency spectra and the predominance of a single haplotype. We suggest that the inactive form of the gene arose in Africa ~100-500 thousand years ago (KYA) and was initially neutral or almost neutral, but that positive selection beginning ~60-100 KYA drove it to near-fixation. We further propose that its selective advantage was sepsis resistance in populations that experienced more infectious diseases as population sizes and densities increased.
Link (pdf)

No comments:
Post a Comment