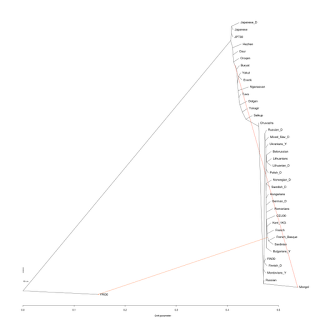
A string of recent papers argued for admixture in human populations at time scales from the Middle Pleistocene to recent centuries. A new paper in Science makes the point convincingly for extensive admixture in humans over the last few thousand years. The authors include the creators of Chromopainter/fineStructure software; the new "Globetrotter" method appears to be a natural extension of that method that seemed to work wonderfully well except for the limitation of producing only a tree of the studied populations.
The paper has a companion website in which you can look up the admixture history of individual populations.
While reading this study, it is important to remember its limitations. Two are immediately obvious: (i) admixture events can only be detected for the last few thousand years, as this method depends on pattern of linkage disequilibrium which decays exponentially with time due to recombination, and (ii) detection of admixture seems to depend on the presence of maximally differentiated populations from the edges of the human geographical range; for example, the Japanese appear unadmixed even though they are clearly of dual Jomon/Yayoi ancestry. On the other hand, the method does detect the admixture present in the San at a similar time scale.
The case of Northwestern Europe appears especially striking as none of the populations from the region show evidence of admixture. This may be because the mixtures taking place there (e.g., between "Celts" and "Anglo-Saxons" in Great Britain) involved populations that were not strongly differentiated. Alternatively, population admixture history may have preceded the last few thousand years and is thus beyond the temporal scope of this method.
An exception to the rule that populations at the edges of the human range appear to be unadmixed are the Armenians who appear to be the only * between the Atlantic and Pacific in Figure 2D (shown at the beginning of this post). The companion site lists their status as "uncertain".
Other results are more questionable; for example, the authors assert that Sardinians are an admixed population with one side being "Egyptian-like" and the other "French-like" whereas the ancient DNA evidence as it stands would rather indicate that Sardinians are the best approximation of Neolithic Europeans currently in existence and so are more likely to (mostly) possess a gene pool that traces back to ~8-9 thousand years in Europe. It will be quite the surprise if so many Europeans from 5kya or earlier look like modern Sardinians and ancient Sardinians don't!
The analysis of Eastern Europe is particularly interesting as it documents three way admixture (Northern/Southern/NE Asian) in most populations but two way admixture (Northern/Southern) in Greeks, estimated at ~37%. The authors claim that this is related to the Slavs, which seems reasonable given the 1,054AD age estimate. On the other hand, according to the companion website, the southern element in Greeks is inferred to be Cypriot-like and it's far from clear that the pre-Slavic population of Greece was Cypriot-like or indeed represented by any of the populations in the authors' dataset.
The three-way admixture in much of eastern Europe is not particularly surprising as history furnishes ample evidence for groups of steppe origin in the region during historical times. Some bequeathed their both language and name (e.g., Magyars), others only their name (e.g., Bulgarians) on the local Europeans, but records indicate a widespread presence of "eastern" groups in Europe from the time of the Huns to that of the Ottomans. A study of late Antique eastern Europeans from the Baltic to the Aegean may help better document how the twin phenomena of the eastern invasions and the spread of the Slavs shaped the present-day genetic diversity of the region.
I suspect that a few ancient samples will be far more informative for understanding the recent history of our species than the most sophisticated modeling of modern populations. Nonetheless, it's great to have a new method that maximizes what can be learned about the past from the messy palimpsest of the present.
Science 14 February 2014:
Vol. 343 no. 6172 pp. 747-751
DOI: 10.1126/science.1243518
A Genetic Atlas of Human Admixture History
Garrett Hellenthal et al.
Modern genetic data combined with appropriate statistical methods have the potential to contribute substantially to our understanding of human history. We have developed an approach that exploits the genomic structure of admixed populations to date and characterize historical mixture events at fine scales. We used this to produce an atlas of worldwide human admixture history, constructed by using genetic data alone and encompassing over 100 events occurring over the past 4000 years. We identified events whose dates and participants suggest they describe genetic impacts of the Mongol empire, Arab slave trade, Bantu expansion, first millennium CE migrations in Eastern Europe, and European colonialism, as well as unrecorded events, revealing admixture to be an almost universal force shaping human populations.
Link
Showing posts with label Mongols. Show all posts
Showing posts with label Mongols. Show all posts
February 13, 2014
July 10, 2013
Population history of middle Euphrates valley
HOMO - Journal of Comparative Human Biology
Available online 3 July 2013
Population history of the middle Euphrates valley: Dental non-metric traits at Tell Ashara, Tell Masaikh and Jebel Mashtale, Syria
Arkadiusz Sołtysiak, Marta Bialon
Fifty-nine dental non-metric traits were scored using Arizona State University Dental Anthropology System on a sample of teeth from 350 human skeletons excavated at three sites in the lower middle Euphrates valley. The dataset was divided into six chronological subsets: Early Bronze Age, Middle Bronze Age, Early Iron Age with Neo-Assyrian period, Classical/Late Antiquity, Early Islamic (Umayyad and Abbasid) period and Modern period. The matrix of Mean Measure of Divergence values exhibited temporal homogeneity of the sample with only dental non-metric trait scores in the Modern subset differing significantly from most other subsets. Such a result suggests that no major gene flow occurred in the middle Euphrates valley between the 3rd millennium BCE and the early 2nd millennium CE. Only after the Mongolian invasion and large depopulation of northern Mesopotamia in the 13th century CE a major population change occurred when the area was taken over in the 17th century by Bedouin tribes from the Arabian Peninsula.
Link
Population history of the middle Euphrates valley: Dental non-metric traits at Tell Ashara, Tell Masaikh and Jebel Mashtale, Syria
Arkadiusz Sołtysiak, Marta Bialon
Fifty-nine dental non-metric traits were scored using Arizona State University Dental Anthropology System on a sample of teeth from 350 human skeletons excavated at three sites in the lower middle Euphrates valley. The dataset was divided into six chronological subsets: Early Bronze Age, Middle Bronze Age, Early Iron Age with Neo-Assyrian period, Classical/Late Antiquity, Early Islamic (Umayyad and Abbasid) period and Modern period. The matrix of Mean Measure of Divergence values exhibited temporal homogeneity of the sample with only dental non-metric trait scores in the Modern subset differing significantly from most other subsets. Such a result suggests that no major gene flow occurred in the middle Euphrates valley between the 3rd millennium BCE and the early 2nd millennium CE. Only after the Mongolian invasion and large depopulation of northern Mesopotamia in the 13th century CE a major population change occurred when the area was taken over in the 17th century by Bedouin tribes from the Arabian Peninsula.
Link
November 03, 2012
Recent admixture in Altaic populations: a legacy of Empire?
Continuing my experiments with ALDER, I took every single Altaic population publicly available, i.e., the following 25 populations:
I then ran ALDER for all 25 Altaic populations using any of the 3*3 West/East Eurasian reference pairs, or a total of 25*3*3= 225 runs. I retained only those 2-ref admixture analyses for which ALDER reported "success" with no warnings.
I then converted reported times to calendar dates: a generation of 29 years was assumed; lacking information about the age of the sampled individuals, I assumed that the "present" is 1980; finally, I report the earliest and latest -/+ limits of any confidence interval, as well as the median of all estimates.
The results can be seen below; for 11 of the 25 populations there was at least one test which was successful with no warnings. This does not mean that the other populations are unadmixed, but the following cases appear to be most "well-behaved":
Now, these appear to make excellent sense.
Of the Dolgans:
Of the Evenks:
Of the Nogais:
Not much needs to be said for the admixture signal in the Uygur, Uzbek, Kyrgyz, and Mongols which collectively ranges from 1260-1500AD. This was a period of Mongol power when Mongolian and Turkic speaking peoples assumed control over Central Asia and replaced to a great degree the previous inhabitants of the area.
The origin of the Balkars is less certain, because they are an old Turkic group that settled in the Caucasus, but the admixture (830-1220AD) date seems plausible. So does, of course, that of the Turks from Caesaria (990-1260AD) which parallels those of my recent experiment, and can be associated with the takeover of Anatolia following the Battle of Manzikert. Finally, I don't have a read explanation for the 11-12th century signal of admixture in the Siberian Altai and Buryat, but presumably it has something to do with the expansions of Altaic peoples around that time that were also felt in the west during this period; presumably, this involved some type of mixture with Caucasoid groups in Siberia.
The admixture dates are quite helpful in helping us better interpret other signals of admixture such as those of ADMIXTURE analyses (e.g., globe13). For example, the Dolgan have 13.1% North_European in that experiment, and the Altai have 13.2%, but apparently this occurred centuries apart and may have involved different groups of West Eurasian people.
In conclusion, ALDER seems to find some quite plausible dates for major admixture episodes in the history of Altaic populations that are compatible with fairly recent historical events.
Altai, Balkars_Y, Buryat, Chuvashs_16, Daur, Dolgan, Evenk_15, Hezhen, Kumyks_Y, Kyrgyz_Bishkek_Ho, Mongol, Mongola, Nogais_Y, Oroqen, Tu, Turkish_Aydin_Ho, Turkish_Istanbul_Ho, Turkish_Kayseri_Ho, Turkmens_Y, Turks, Tuva, Uygur, Uzbeks, Xibo, YakutI also took three West Eurasian populations unlikely to have historical East Asian admixture (French, French_Basque, and Sardinians), and three East Eurasian populations unlikely to have historical West Eurasian admixture (Dai, She, Miaozu). I merged all of the above in PLINK with a --geno 0.03 flag, and extracting SNPs present in the Rutgers recombination map for Illumina chips (a total of 524,822 SNPs).
I then ran ALDER for all 25 Altaic populations using any of the 3*3 West/East Eurasian reference pairs, or a total of 25*3*3= 225 runs. I retained only those 2-ref admixture analyses for which ALDER reported "success" with no warnings.
I then converted reported times to calendar dates: a generation of 29 years was assumed; lacking information about the age of the sampled individuals, I assumed that the "present" is 1980; finally, I report the earliest and latest -/+ limits of any confidence interval, as well as the median of all estimates.
The results can be seen below; for 11 of the 25 populations there was at least one test which was successful with no warnings. This does not mean that the other populations are unadmixed, but the following cases appear to be most "well-behaved":
Now, these appear to make excellent sense.
Of the Dolgans:
There also existed a group of Russian settlers on the River Heta, who, by the end of the 19th century, had become Dolganized and had gradually adopted the way of life of nomadic reindeer breeders. ... The tribes forming the nucleus of the Dolgans migrated from the banks of the River Lena at the end of the 17th century. One of the reasons for migration was the fact that Russian goods, flour, for instance, were coming to the Taimyr Peninsula by the boats on the Lena.The 1770-1860AD range for the admixture appears to coincide with the period where the Dolgans came under Russian influence.
Of the Evenks:
The history of the Evenks' habitation can be traced in detail from the 17th century on. At that time the Evenks left several of their previous territories, for instance, the River Angara, when the Yakut, the Buryat and the Russians appeared in the province. The Evenks had especially bad relations with the Yakuts, who had settled in the river basin of the Lena in the 13th century. In the 18th and 19th centuries the Evenks living there adopted the Yakut language. In the Baikal area the Evenks began to speak the Buryat and the Mongolian languages, and even converted to lamaism. The southern Evenk -- the Manegir, the Birar, the Solon -- were influenced by the Manchu, Daur and Chinese cultures. The arable lands in Siberia were occupied by Russian settlers, migrating there in the 17th century, and those Evenks, living in the vicinity on the upper reaches of the Lena and near Baikal, were russified.Again, the 1630-1800AD admixture range seems consistent with the time when Evenks came into contact with Russians.
Of the Nogais:
In the first half of the 17th century a number of Nogay tribes were nomadic on the steppes between the Danube and the Caspian. The invasion of the warlike Kalmyks forced several of the Nogay tribes to leave their home steppes and withdraw to the foothills of the North Caucasus. By the River Kuban they met with the Cherkess. In the Moscow chronicles from the 16th and 17th centuries there are several mentions of the Nogay, including the two Nogay Hordes, the Great and the Small. The former roamed beyond the River Volga, the latter somewhat to the west. Both had numerous military encounters with the Russians. In the 17th century some of the Nogay chiefs entered into an alliance with Moscow and fought at times together with the Russians against the Kabardians, the Kalmyks and peoples of Dagestan.The 1610-1730AD range intersects the period when the Nogais settled in the North Caucasus and interacted with North Caucasians and Russians.
Not much needs to be said for the admixture signal in the Uygur, Uzbek, Kyrgyz, and Mongols which collectively ranges from 1260-1500AD. This was a period of Mongol power when Mongolian and Turkic speaking peoples assumed control over Central Asia and replaced to a great degree the previous inhabitants of the area.
The origin of the Balkars is less certain, because they are an old Turkic group that settled in the Caucasus, but the admixture (830-1220AD) date seems plausible. So does, of course, that of the Turks from Caesaria (990-1260AD) which parallels those of my recent experiment, and can be associated with the takeover of Anatolia following the Battle of Manzikert. Finally, I don't have a read explanation for the 11-12th century signal of admixture in the Siberian Altai and Buryat, but presumably it has something to do with the expansions of Altaic peoples around that time that were also felt in the west during this period; presumably, this involved some type of mixture with Caucasoid groups in Siberia.
The admixture dates are quite helpful in helping us better interpret other signals of admixture such as those of ADMIXTURE analyses (e.g., globe13). For example, the Dolgan have 13.1% North_European in that experiment, and the Altai have 13.2%, but apparently this occurred centuries apart and may have involved different groups of West Eurasian people.
In conclusion, ALDER seems to find some quite plausible dates for major admixture episodes in the history of Altaic populations that are compatible with fairly recent historical events.
July 22, 2012
Clarifying the phylogeny of Y-chromosome haplogroup C3c
A short and to the point paper that addresses the issue of classification within Y-haplogroup C3c and refines our knowledge about the distribution of both C3c* and C3c1. I wish more researchers would publish such short technical papers that refine the classification of their Y-chromosome samples as more phylogenetic information becomes available.
From the paper:
J Hum Genet. 2012 Jul 19. doi: 10.1038/jhg.2012.93. [Epub ahead of print]
On the Y-chromosome haplogroup C3c classification.
Malyarchuk BA, Derenko M, Denisova G.
Abstract As there are ambiguities in classification of the Y-chromosome haplogroup C3c, relatively frequent in populations of Northern Asia, we analyzed all three haplogroup-defining markers M48, M77 and M86 in C3-M217-individuals from Siberia, Eastern Asia and Eastern Europe. We have found that haplogroup C3c is characterized by the derived state at M48, whereas mutations at both M77 and M86 define subhaplogroup C3c1. The branch defined by M48 alone would belong to subhaplogroup C3c*, characteristic for some populations of Central and Eastern Siberia, such as Koryaks, Evens, Evenks and Yukaghirs. Subhaplogroup C3c* individuals could be considered as remnants of the Neolithic population of Siberia, based on the age of C3c*-short tandem repeat variation amounting to 4.5±2.4 thousand years.
Link
From the paper:
In our study, the highest frequencies of subhaplogroup C3c1-(M77, M86) were observed in Tungusic-speaking people of North-Eastern Asia, such as Evens and Evenks, as well as in Turkic-speaking Altaian Kazakhs and Mongolic-speaking Kalmyks. These results are in agreement with previous observations based on separate or joint genotyping of M77 and M86 markers.3,9,12,16The authors apply the evolutionary mutation rate -although they acknowledge that molecular dating is controversial- to obtain ages of 9.9 (C3c), 6.5 (C3c1), and 4.5 (C3c*). While I don't trust the ability of Y-STR-based molecular dating to provide reasonably accurate age estimates, I would not be surprised if C3c1 was somehow implicated in the deeper origins of the Altaic language family, at least in the "narrow-sense" (Mongolian-Tungusic-Turkic).
C3c* haplotypes were detected in aboriginal populations of North- Eastern Asia—Koryaks (28.2%) and Evens (1.6%) from the Sea of Okhotsk coast (Magadan region) and West Evenks (2.4%) from Central Siberia (Evenki Autonomous District) (Table 1). Earlier, two Evenk individuals from southern part of Yakutia, one Yakut-speaking Evenk and one Yukaghir were found to belong to C3c*.2,3 Therefore, the geographic distribution of subhaplogroup C3c* is limited to the eastern part of Siberia.
J Hum Genet. 2012 Jul 19. doi: 10.1038/jhg.2012.93. [Epub ahead of print]
On the Y-chromosome haplogroup C3c classification.
Malyarchuk BA, Derenko M, Denisova G.
Abstract As there are ambiguities in classification of the Y-chromosome haplogroup C3c, relatively frequent in populations of Northern Asia, we analyzed all three haplogroup-defining markers M48, M77 and M86 in C3-M217-individuals from Siberia, Eastern Asia and Eastern Europe. We have found that haplogroup C3c is characterized by the derived state at M48, whereas mutations at both M77 and M86 define subhaplogroup C3c1. The branch defined by M48 alone would belong to subhaplogroup C3c*, characteristic for some populations of Central and Eastern Siberia, such as Koryaks, Evens, Evenks and Yukaghirs. Subhaplogroup C3c* individuals could be considered as remnants of the Neolithic population of Siberia, based on the age of C3c*-short tandem repeat variation amounting to 4.5±2.4 thousand years.
Link
March 31, 2012
Three quarters of Kerey clan men belong to Genghis Khan Y chromosome cluster
From the paper:
More:
Human Biology: Vol. 84: Iss. 1, Article 4.
The Y-chromosome C3* star-cluster attributed to Genghis Khan's descendants is present at high frequency in the Kerey clan from Kazakhstan
Serikbai Abilev et al.
In order to verify the possibility that the Y-chromosome C3* star-cluster attributed to Genghis Khan and his patrilineal descendants is relatively frequent in the Kereys, who are the dominant clan in Kazakhstan and in Central Asia as a whole, polymorphism of the Y-chromosome was studied in Kazakhs, represented mostly by members of the Kerey clan. The Kereys showed the highest frequency (76.5%) of individuals carrying the Y-chromosome variant known as C3* star-cluster ascribed to the descendants of Genghis Khan. C3* star-cluster haplotypes were found in two sub-clans, Abakh-Kereys and Ashmaily-Kereys, diverged about 20-22 generations ago according to the historical data. Median network of the Kerey star-cluster haplotypes at 17 STR loci displays a bipartite structure, with two subclusters defined by the only difference at DYS448 locus. It is noteworthy that there is a strong correspondence of these subclusters with the Kerey sub-clans affiliation. The data obtained suggest that the Kerey clan appears to be the largest known clan in the world descending from a common Y-chromosome ancestor. Possible ways of Genghis Khan‟s relation to the Kereys are discussed.
Link
According to the historical data, the split between two sub-clans of the Kereys occurred about 20-22 generations ago (Khalidullin 2005). Estimation of divergence time (TD) of two groups of 15 STR haplotypes (except for DYS385a,b loci) found in the Kereys sub-clans demonstrates that TD value equal to 630 ± 190 years (or approximately 21 ± 6 generations) is resulted when a mean of per-locus, per-generation mutation rate of 0.0033 and a 30-year generation time are used. Note that similar value of mutation rate (0.00324) has been calculated as optimal for 15 STR haplotypes by Busby et al. (2011) who have investigated the question on how average squared distance (ASD) estimates change within haplotype sets when using different combinations of Y-chromosome STRs. This mutation rate belongs to a class of so called genealogical STR mutation rates revealed by direct observation in father/son pairs (Kayser et al. 2000; Goedbloed et al. 2009).The correspondence between the split time of the Kerey sub-clans and the age estimate of their Y-STR divergence is quite interesting and provides an independent historical argument for the correspondence between the C3* star cluster and Genghis Khan (or at least his direct patrilineal kin). Note that the star cluster's age matches G. K. only using a genealogical mutation rate, and not the widely (mis)used "effective mutation rate. The timeframe is recent enough to render any saturation effects from non-linearity (as described by Busby et al.) relatively unimportant.
More:
The data reported above, taken together with the known arguments in favor of the
possible Genghis Khan‟s descent of Y-chromosome C3* star-cluster (Zerjal et al. 2003), allow us to suggest two hypotheses.
(1) The star-cluster is not directly related to the descendants of Genghis Khan, but rather is associated with the Kerait clan members. Mongol conquest with participation of the Keraits as special Khan‟s military forces allowed them to disseminate the Kerait-specific Y-chromosomes in the vast area inhabited by various peoples.
(2) Genghis Khan by himself belonged to the Keraits. This is supported by the following historical evidence (Man 2004; Khalidullin 2005). The Keraits inhabited the banks of the Onon River, where the camp of Genghis Khan‟s father Yesukhei was located. Yesukhei was declared as a blood brother of the Keraits‟ Khan Toghrul (Wang Khan). Toghrul then declared Genghis Khan his son-in-law. Fraternization of the Genghis Khan family with the Keraits‟ Khan suggests that a real blood relationship, though probably not approved officially, existed between them.
Human Biology: Vol. 84: Iss. 1, Article 4.
The Y-chromosome C3* star-cluster attributed to Genghis Khan's descendants is present at high frequency in the Kerey clan from Kazakhstan
Serikbai Abilev et al.
In order to verify the possibility that the Y-chromosome C3* star-cluster attributed to Genghis Khan and his patrilineal descendants is relatively frequent in the Kereys, who are the dominant clan in Kazakhstan and in Central Asia as a whole, polymorphism of the Y-chromosome was studied in Kazakhs, represented mostly by members of the Kerey clan. The Kereys showed the highest frequency (76.5%) of individuals carrying the Y-chromosome variant known as C3* star-cluster ascribed to the descendants of Genghis Khan. C3* star-cluster haplotypes were found in two sub-clans, Abakh-Kereys and Ashmaily-Kereys, diverged about 20-22 generations ago according to the historical data. Median network of the Kerey star-cluster haplotypes at 17 STR loci displays a bipartite structure, with two subclusters defined by the only difference at DYS448 locus. It is noteworthy that there is a strong correspondence of these subclusters with the Kerey sub-clans affiliation. The data obtained suggest that the Kerey clan appears to be the largest known clan in the world descending from a common Y-chromosome ancestor. Possible ways of Genghis Khan‟s relation to the Kereys are discussed.
Link
March 16, 2012
TreeMix analysis of North Eurasians (and an African surprise)
I have used my K12b dataset to isolate a set of 537 individuals who had less than 10% membership in the South Asian, Northwest African, Southeast Asian, South Asian, East African, Gedrosia, South Asian, East African, Southwest_Asian, and Sub_Saharan components. Hence, the remaining 537 individuals had 90%+ membership in the remaining Atlantic_Med, North_European, Caucasus, Siberian, and East_Asian components.
- The Atlantic_Med component is frequent in northwestern Europe
- The North_European component is dominant in northeastern Europe and forays into Siberia
- The Caucasus component is dominant in the Caucasus and forays into Central Asia
- The Siberian component is dominant in North Asia and forays into Europe
- The East_Asian component is frequent in East Asia and forays into North Asia
Russian_D, Polish_D, German_D, Finnish_D, Swedish_D, Mixed_Slav_D, Norwegian_D, Lithuanian_D, Japanese_D, Daur, French, French_Basque, Hezhen, Japanese, Oroqen, Russian, Sardinian, Yakut, CEU30, JPT30, Belorussian, Chuvashs, Hungarians, Lithuanians, Romanians, Selkup, Evenk, Tuva, Yukagir, Nganassan, Dolgan, Buryat, Mongol, FIN30, Kent_1KG, Bulgarians_Y, Ukranians_Y, Mordovians_Y
Additionally, a sample of 30 Yoruba from the HapMap-3 was used as an outgroup.
TreeMix analysis
The TreeMix analysis was performed with default parameters, and allowing for a different number of migration edges.
Nomenclature: The direction of gene flow is best seen in the figure and/or associated treeout files.
For the text, I will put in (), the common ancestor of two populations, e.g., (French_Basque,Sardinian) and also as (X, *) the tree rooted at a particular node X, e.g., (Buryat, *)
0 migration edges:
The West and East Eurasian clusters are identified, with some populations with likely admixture being placed closer to the Eurasian root.
1 migration edge:
64% from (Sardinians/Basques) to Yoruba; this is difficult to interpret, but there has been evidence in the past that Africans and West Eurasians share more ancestry than Africans and East Asians do. In the linked post, I proposed a major episode of back-migration into Africa, and it is perhaps this that is being captured by this migration edge: Sardinians/Basques are the only two South-West Eurasian populations included, and any back-migration into Africa must have originated in the southern parts of West Eurasia.
Such a high level of back-migration may in fact be plausible, since Yoruba are a predominantly Y-haplogroup E bearing population, and the origin of the DE clade of the human Y-chromosome phylogeny is up in the air with both an African and Eurasian case having been advanced. Personally, I favor the Eurasian case, since within the CT clade, we have two subclades: CF (Eurasian) and DE (Eurasian/African).
Such a high level of back-migration may in fact be plausible, since Yoruba are a predominantly Y-haplogroup E bearing population, and the origin of the DE clade of the human Y-chromosome phylogeny is up in the air with both an African and Eurasian case having been advanced. Personally, I favor the Eurasian case, since within the CT clade, we have two subclades: CF (Eurasian) and DE (Eurasian/African).
Interestingly, John Hawks has recently discovered an unanticipated excess of "Neandertal ancestry" in Yoruba. This may also point to a back-migration into Africa and/or admixture of a group of Africans related to Eurasians (whom I've called Afrasians), with groups of Africans (Palaeoafricans) that split before the H. sapiens/H. neandertalensis common ancestor.
There is, however, another detail in the figure that may have escaped your notice: there is now about 0.5 worth of drift in the figure (left-to-right) as opposed to only 0.12 in the tree without migration edges. So, perhaps what we are seeing is indeed the first sign of admixture between modern and archaic humans in Africa, which has been made more likely by recent anthropological discoveries.
It's not clear to me whether TreeMix has stumbled onto something important or not, but it is certainly worth keeping in mind that the above model fits the data better than the simple tree model. Moreover, TreeMix attempts to reverse the polarity of migration edges, and -apparently- the (Sardinian, French_Basque)-to-Yoruba edge is preferable to the reverse.
So, we should keep our minds open to the possibility that the greater similarity of West Eurasians to Africans is not the result of multiple Out-of-Africa waves, one of which affected only West Eurasians, but of an Into-Africa back-migration from West Eurasia.
So far, tree-based models have focused on how diverse African groups are, and hence, the reduced diversity of Eurasians has been interpreted as an Out-of-Africa bottleneck that carried a subset of African variation into Eurasia.
But, there is an alternative interpretation of the evidence, namely that African groups are diverse because they carry a superset of ancient Into-Africa variation, with the African-specific part of their variation being the result of admixture with pre-existing African hominins. Such a scenario cannot be captured by tree models, but is apparently considered and not rejected by TreeMix which allows for lateral gene flow. Let's wait and see what new things come from full genome sequencing.
So, we should keep our minds open to the possibility that the greater similarity of West Eurasians to Africans is not the result of multiple Out-of-Africa waves, one of which affected only West Eurasians, but of an Into-Africa back-migration from West Eurasia.
So far, tree-based models have focused on how diverse African groups are, and hence, the reduced diversity of Eurasians has been interpreted as an Out-of-Africa bottleneck that carried a subset of African variation into Eurasia.
But, there is an alternative interpretation of the evidence, namely that African groups are diverse because they carry a superset of ancient Into-Africa variation, with the African-specific part of their variation being the result of admixture with pre-existing African hominins. Such a scenario cannot be captured by tree models, but is apparently considered and not rejected by TreeMix which allows for lateral gene flow. Let's wait and see what new things come from full genome sequencing.
2 migration edges:
The (French_Basque/Sardinian)-to-Yoruba edge persists (64%) and a new edge was added from (Buryat, *)-to-Mongol (85%). The "Mongol" sample consists of Siberian Mongols described by Rasmussen et al. (2010). An inspection of their K12b population portrait indicates that they do, in fact, have West Eurasian admixture, which according to the K12b spreadsheet amounts to about 18% in total.
3 migration edges:
The aforementioned (French_Basque/Sardinian)-to-Yoruba (64%) and (Buryat,*)-toMongol (85%) edges persist, and now we have a 68% Nganasan-to-Selkup edge.
These are the two Siberian Uralic populations in the dataset. This seems to parallel the K12b results, as Selkups have a North_European element which the Nganasans (Uralic speakers from the Arctic coast of Central Siberia lack), so we are seeing the hybridity of the Selkups here, who, like the Mongol sample are partly of West Eurasian ancestry.
4 migration edges:
The aforementioned (French_Basque,Sardinian)-to-Yoruba (64%), (Buryat,*)-to-Mongol (84%), and Nganasan-to-Selkup (68%) persist, and now we have a 89% (Buryat, *)-to-Tuva edge. According to the K12b the Tuva have 13.3% West Eurasian admixture, so again we have reasonably good agreement between TreeMix and ADMIXTURE.
Interestingly, the non-"eastern" component of Selkups and Tuvans now forms a clade. It seems that a Nganasan-like and a (Buryat, *)-like population have converged into southern Siberia, absorbed a common local element and became the Selkup and Tuva respectively.
5 migration edges:
The aforementioned (French_Basque,Sardinian)-to-Yoruba (64%), (Buryat,*)-to-Mongol (85%), Nganasan-to-Selkup (68%) persist, and 90% (Buryat, *)-to-Tuva persist, and now we have a new 18% Oroqen-to-(Yakut, Evenk) edge. The Oroqen and the Evenk are Tungusic speakers, whereas the Yakut are Turkic people from northeastern Siberia, having migrated there from the vicinity of Lake Baikal during the last millennium.
6 migration edges:
6 migration edges:
The aforementioned (French_Basque,Sardinian)-to-Yoruba (64%), (Buryat,*)-to-Mongol (85%), Nganasan-to-Selkup (68%), 90% (Buryat, *)-to-Tuva persist, 18% Oroqen-to-(Yakut, Evenk), persist, and a new 16% Nganasan-to-Oroqen edge appears. Interestingly, this has allowed the Oroqen and Hezhen to now form their own clade, which makes sense as these are both Tungusic speakers from northeastern China. The other Tungusic population, the Evenk group with the Turkic Yakut: what they share in common is that they both share origins close to Lake Baikal in Siberia.
7 migration edges:
The aforementioned (French_Basque,Sardinian)-to-Yoruba (64%), (Buryat,*)-to-Mongol (85%), Nganasan-to-Selkup (68%), 90% (Buryat, *)-to-Tuva persist, 18% Oroqen-to-(Yakut, Evenk), 16% Nganasan-to-Oroqen edges persist, and there is a new 81% Evenk-to-Yukagir edge. The remainder of the Yukagirs' ancestry is derived from the West Eurasian tree. The Yukagir language is rather mysterious, with some links to Uralic having been postulated. Here it pays off to look at the population portraits, since it is apparent that -unlike the Selkup- their West Eurasian ancestry is limited to a few individuals.
It is fairly interesting that Russian anthropologists placed the Yukagirs in the Baikal group of the Central Asian race, the same as the Evenks, who are their biggest donors. So, Yakuts, Evenks, and Yukagirs all seem to share the same Baikal-type of origin.
8 migration edges:
7 migration edges:
The aforementioned (French_Basque,Sardinian)-to-Yoruba (64%), (Buryat,*)-to-Mongol (85%), Nganasan-to-Selkup (68%), 90% (Buryat, *)-to-Tuva persist, 18% Oroqen-to-(Yakut, Evenk), 16% Nganasan-to-Oroqen edges persist, and there is a new 81% Evenk-to-Yukagir edge. The remainder of the Yukagirs' ancestry is derived from the West Eurasian tree. The Yukagir language is rather mysterious, with some links to Uralic having been postulated. Here it pays off to look at the population portraits, since it is apparent that -unlike the Selkup- their West Eurasian ancestry is limited to a few individuals.
It is fairly interesting that Russian anthropologists placed the Yukagirs in the Baikal group of the Central Asian race, the same as the Evenks, who are their biggest donors. So, Yakuts, Evenks, and Yukagirs all seem to share the same Baikal-type of origin.
8 migration edges:
There is now a 64% Sardinian-to-Yoruba edge, a 16% Oroqen-to-Yukagir edge, 20% (Buryat, *)-to-(Yakut, Evenk), and a 24% Nganasan-to-Chuvash edge, 29% Oroqen-to-(Yakut,Evenk) edge, 88% (Buryat, *)-to-Tuva, 62% Nganasan-to-Selkup, 85% (Buryat, *)-to-Mongol.
The tree has been rather re-organized, with two main Siberian groups identified: an eastern group (Hezhen, Daur, Oroqen, Buryat), and a central group (Yukagir, Dolgan, Nganasan, Yakut, Evenk, Selkup). The Chuvash, predominantly Europeoid Turkic speakers from Russia show evidence of gene flow from the central group as well, whereas the Selkup, Uralic speakers from Siberia, who belong to the central group, show evidence of gene flow from Europe.
9 migration edges:
64% (French_Basque,Sardinian)-to-Yoruba, 85% (Buryat, *)-to-Mongol, 68% Nganasan-to-Selkup, 92% (Buryat,*)-to-Tuva, 14% Oroqen-to-(Yakut,Evenk), 14% Nganasan-to-Oroqen, 82% Yakut-to-Yukagir, 90% Evenk-to-Dolgan, 13% Hezhen-to-(Nganasan, *).
10 migration edges:
64% (French_Basque, Sardinian)-to-Yoruba, (85% Nganasan, *)-to-Mongol, 68% Nganasan-to-Selkup, 92% (Nganasan,*)-to-Tuva, 15% Oroqen-to-(Yakut,Evenk), 15% Nganasan-to-Oroqen, 82% Yakut-to-Yukagir, 90% Evenk-to-Dolgan, 43% Hezhen-to-Buryat, 14% Sardinian-to-Bulgarian.
I will stop at this point. I may add more migration edges later to this post, but I'm tired of typing this stuff.
You can download all the plots and *.treeout files here.
UPDATE (March 20): I have repeated the experiment with HGDP San, rather than Yoruba as the outrgroup:
There is now a 63% migration edge from (Basque, Sardinian) to San.
UPDATE (March 20): I have repeated the experiment with HGDP San, rather than Yoruba as the outrgroup:
There is now a 63% migration edge from (Basque, Sardinian) to San.
December 28, 2011
Genetic structure in China
After my experiment on Spain, I decided to carry out a similar experiment in China, for which there is a large number of regional/ethnic sub-populations.15 clusters were inferred with 22 MDS dimensions.
The Uygur are the clear outlier population, doubtlessly due to their substantial Caucasoid admixture and geographical position in Central Asia, a region that was traditionally at the outskirts of Chinese civilization. Other Altaic speakers (both Mongolic and Tungusic) are also divergent, as are the Dai/Lahu people from the China/Thailand/Laos area.
Interestingly, the Tujia people from Central China seem to be the ones most like the Han overall, with Hmongic Miaozu/She more like the southern Han.
The Uygur are the clear outlier population, doubtlessly due to their substantial Caucasoid admixture and geographical position in Central Asia, a region that was traditionally at the outskirts of Chinese civilization. Other Altaic speakers (both Mongolic and Tungusic) are also divergent, as are the Dai/Lahu people from the China/Thailand/Laos area.
Interestingly, the Tujia people from Central China seem to be the ones most like the Han overall, with Hmongic Miaozu/She more like the southern Han.
May 24, 2011
The reality of the Altaic language family
 Personally I'm not surprised by this; my own look at genomic data has identified an "Altaic" component which peaks at the Turkic Yakut and Tungusic Evenk, and is shared by every Turkic, Mongolic, and Tungusic population available to me. The same component also occurs to some extent among all the Japanese (5) and Korean (4) members of the Dodecad Project, while it is lacking in all the Chinese ones (8).
Personally I'm not surprised by this; my own look at genomic data has identified an "Altaic" component which peaks at the Turkic Yakut and Tungusic Evenk, and is shared by every Turkic, Mongolic, and Tungusic population available to me. The same component also occurs to some extent among all the Japanese (5) and Korean (4) members of the Dodecad Project, while it is lacking in all the Chinese ones (8).
Of particular interest is the degree of CCM between Indo-European and Semitic languages (Tables 2 and 3). In many of the most geographically distant languages these are less than 10; by comparison, among Semitic languages the are all greater than 20. This seems to be quite in agreement with the idea that Semitic is a Bronze Age language family, Indo-European a Neolithic one.
This impression is strengthened by the fact that CCM between reconstructed proto-languages (e.g. Proto-Iranian and Proto-Slavic = 20) are much higher. Since these proto-languages are a few thousand years closer to the root of PIE than present-day languages, and differences between them are similar to those of Semitic languages, the notion that PIE is a few thousand years older than Proto-Semitic seems quite consistent with the evidence.
Journal of Language Relationship • Вопросы языкового родства • 3 (2010) • Pp. 117–126 • © Turchin P., Peiros I., Gell-Mann M., 2010
Analyzing genetic connections between languages by matching consonant classes
Peter Turchin (University of Connecticut)
Ilia Peiros (Santa Fe Institute)
Murray Gell-Mann (Santa Fe Institute)
Ilia Peiros (Santa Fe Institute)
Murray Gell-Mann (Santa Fe Institute)
The idea that the Turkic, Mongolian, Tungusic, Korean, and Japanese languages are genetically related (the “Altaic hypothesis”) remains controversial within the linguistic community. In an effort to resolve such controversies, we propose a simple approach to analyzing genetic connections between languages. The Consonant Class Matching (CCM) method uses strict phonological identification and permits no changes in meanings. This allows us to estimate the probability that the observed similarities between a pair (or more) of languages occurred by chance alone. The CCM procedure yields reliable statistical inferences about historical connections between languages: it classifies languages correctly for well-known families (Indo-European and Semitic) and does not appear to yield false positives. The quantitative patterns of similarity that we document for languages within the Altaic family are similar to those in the non-controversial Indo-European family. Thus, if the Indo-European family is accepted as real, the same conclusion should also apply to the Altaic family.
Link (pdf)
Link (pdf)
March 14, 2011
Y chromosomes of Altaian Kazakhs
This paper uses both the pedigree (genealogical or germline) and evolutionary mutation rates. Readers of the blog are aware that I've been a vehement critic of the latter on theoretical grounds since 2008, and I've started keeping track of cases where the germline rate has a better fit to the archaeological record than the evolutionary rate.
The younger ages of the Mongoloid lineages in this population makes good historical sense, as these are derived from tribal Turko-Mongolian tribes establishing (more recently) control over the pre-existing Iranian populations of the steppe. The gene pool of the latter has been marginalized but it maintains its genetic diversity.
The presence of haplogroup J2a here as the modal Caucasoid lineage, followed by haplogroups G1 and G2a is also quite interesting, and plausibly brings origin of the ancestors of the pre-Altaic inhabitants of the region in close proximity to the West Asian homeland of the ancestors of the Indo-Aryans.
PLoS ONE 6(3): e17548. doi:10.1371/journal.pone.0017548
Y-Chromosome Variation in Altaian Kazakhs Reveals a Common Paternal Gene Pool for Kazakhs and the Influence of Mongolian Expansions
Matthew C. Dulik et al.
Kazakh populations have traditionally lived as nomadic pastoralists that seasonally migrate across the steppe and surrounding mountain ranges in Kazakhstan and southern Siberia. To clarify their population history from a paternal perspective, we analyzed the non-recombining portion of the Y-chromosome from Kazakh populations living in southern Altai Republic, Russia, using a high-resolution analysis of 60 biallelic markers and 17 STRs. We noted distinct differences in the patterns of genetic variation between maternal and paternal genetic systems in the Altaian Kazakhs. While they possess a variety of East and West Eurasian mtDNA haplogroups, only three East Eurasian paternal haplogroups appear at significant frequencies (C3*, C3c and O3a3c*). In addition, the Y-STR data revealed low genetic diversity within these lineages. Analysis of the combined biallelic and STR data also demonstrated genetic differences among Kazakh populations from across Central Asia. The observed differences between Altaian Kazakhs and indigenous Kazakhs were not the result of admixture between Altaian Kazakhs and indigenous Altaians. Overall, the shared paternal ancestry of Kazakhs differentiates them from other Central Asian populations. In addition, all of them showed evidence of genetic influence by the 13th century CE Mongol Empire. Ultimately, the social and cultural traditions of the Kazakhs shaped their current pattern of genetic variation.
Link
In this particular case, the difference between the two rates (about 3-fold) is especially interesting, because of the whole "Genghis Khan" theory according to which a large number of central Asian men belong to a haplotype cluster dated to around the time of the Mongol conqueror and maybe the descendants of Genghis and his close male relatives. This theory relies on the use of the germline rate: otherwise the genetic signature attributed to the Khan must be redated to a much earlier time.
The authors give a convincing argument in favor of the pedigree rate:
The difficulty in reliably determining the coalescent dates for the lineages found in Kazakh populations makes it nearly impossible to determine whether these lineages were present in ancestral nomadic steppe groups (Scythians, Xiongnu, Xianbei, Toba, and Jou-Jan) or were contributed by the descendents of Genghis Khan and the Mongol armies that, at one time, held control over the region. An important reason for caution here is the current debate about the most appropriate mutation rate for NRY coalescence estimates. The evidence provided by Zerjal et al. [14] supports the younger estimates, suggesting that the Kazakh haplotypes could be the direct result of the Mongol influence in the 13th century CE. The presence of the C3* haplotype cluster in the Kazakh also supports the genealogical assertions that (for at least some Kazakh men) there is a direct paternal connection to Genghis Khan.If the evolutionary rate is the more accurate value for Y-STRs, then the Kazakh lineages coalesce to roughly 2,000 years ago. This date suggests a far older source for them, possibly with the westward movements of Altaic-speaking peoples around the second and first centuries BCE. In this case, we would expect to see multiple haplotype clusters exhibiting a similar pattern as the Genghis Khan cluster. However, we do not observe this pattern. As Zerjal et al. [14] pointed out, this haplotype cluster is unique. Therefore, given the evidence presented here and in Zerjal et al. [14], we believe the best interpretation of the data is that Kazakh Y-chromosome diversity was strongly influenced by the Mongols of the 13th century CE.
The younger ages of the Mongoloid lineages in this population makes good historical sense, as these are derived from tribal Turko-Mongolian tribes establishing (more recently) control over the pre-existing Iranian populations of the steppe. The gene pool of the latter has been marginalized but it maintains its genetic diversity.
The presence of haplogroup J2a here as the modal Caucasoid lineage, followed by haplogroups G1 and G2a is also quite interesting, and plausibly brings origin of the ancestors of the pre-Altaic inhabitants of the region in close proximity to the West Asian homeland of the ancestors of the Indo-Aryans.
PLoS ONE 6(3): e17548. doi:10.1371/journal.pone.0017548
Y-Chromosome Variation in Altaian Kazakhs Reveals a Common Paternal Gene Pool for Kazakhs and the Influence of Mongolian Expansions
Matthew C. Dulik et al.
Kazakh populations have traditionally lived as nomadic pastoralists that seasonally migrate across the steppe and surrounding mountain ranges in Kazakhstan and southern Siberia. To clarify their population history from a paternal perspective, we analyzed the non-recombining portion of the Y-chromosome from Kazakh populations living in southern Altai Republic, Russia, using a high-resolution analysis of 60 biallelic markers and 17 STRs. We noted distinct differences in the patterns of genetic variation between maternal and paternal genetic systems in the Altaian Kazakhs. While they possess a variety of East and West Eurasian mtDNA haplogroups, only three East Eurasian paternal haplogroups appear at significant frequencies (C3*, C3c and O3a3c*). In addition, the Y-STR data revealed low genetic diversity within these lineages. Analysis of the combined biallelic and STR data also demonstrated genetic differences among Kazakh populations from across Central Asia. The observed differences between Altaian Kazakhs and indigenous Kazakhs were not the result of admixture between Altaian Kazakhs and indigenous Altaians. Overall, the shared paternal ancestry of Kazakhs differentiates them from other Central Asian populations. In addition, all of them showed evidence of genetic influence by the 13th century CE Mongol Empire. Ultimately, the social and cultural traditions of the Kazakhs shaped their current pattern of genetic variation.
Link
November 07, 2010
Multidimensional scaling and ADMIXTURE across Northern Eurasia corresponds to geography and language
 Here is a multi-dimensional scaling plot of a number of North Eurasian populations. In comparison to my previous post, I have excluded Americans and Greenlanders, and added several other populations from Central Asia and West Eurasia.
Here is a multi-dimensional scaling plot of a number of North Eurasian populations. In comparison to my previous post, I have excluded Americans and Greenlanders, and added several other populations from Central Asia and West Eurasia. Population labels have been printed in the co-ordinates of the population averages; these largely correspond with identifiable blobs of colored points, but note that some populations have several outliers, so labels appear in white space. Most notable in that respect are the Koryak, Chukchi, and the Nganasan, all of whom have some apparently European-admixed individuals.

"Mongol" corresponds to Rasmussen et al. (2010) Mongol sample, while "Mongola" to the HGDP-CEPH one. The population codes on the left may not be clearly visible as they overlap with each other and are CEU, LT, HU (relatively unadmixed Caucasoids), FI/RU (Uralian-admixed northern Caucasoids), IR/TR (Altaic-admixed southern Caucasoids). The West Eurasian part of the plot can be seen blown up on the right.
The correspondence with geography and language is striking. Siberian isolates from the extreme north and east, Koryak and Chuckhi are on top; HapMap Chinese at the bottom. Between them are Uralians (Selkup, Yukagir, Nganassan) and Altaics (Mongol-Tungus-Turkic people).
Below is ADMIXTURE analysis for the same set of populations, for K=7:
The close relationship between the two Caucasoid components is apparent (Fst=0.033), but note fairly large Fst divergences between the morphologically Mongoloid groups. I attribute this mostly to the very low population sizes of these groups, which have probably affected them by drift. For the less demographically constrained Altaic and East Asian components, Fst=0.044.
 If you are not familiar with these ethnic groups, the Red Book of the Peoples of the Russian Empire and the Ethnologue indexes on Altaic and Uralic are invaluable, as are the portraits of ethnic groups of China. On the right a picture of a Nganasan.
If you are not familiar with these ethnic groups, the Red Book of the Peoples of the Russian Empire and the Ethnologue indexes on Altaic and Uralic are invaluable, as are the portraits of ethnic groups of China. On the right a picture of a Nganasan.UPDATE: Also, a past post from the blog, collating Y-haplogroup N frequencies with anthropological descriptions. Nganasans apparently belong to haplogroup N at a frequency of 92.1%!
November 06, 2010
ADMIXTURE in Siberia, Greenland, and Alaska
I have discovered a great dataset from Rasmussen et al. (2010). The data had been used before in conjunction with an ancient DNA sequence, but for me it is invaluable, as it fills up one of the major holes in Eurasia, namely Siberia, and includes a number of Altaic, Uralic, and other North Eurasian people. I suspect that this will be invaluable in fine-tuning the Northeast Asian ancestry of Dodecad Project members.
To begin with, after I processed the data, I ran ADMIXTURE on it up to K=7. Below you can see the results for K=7:
I'm no expert in linguistics, but it's clear to me that the light blue component corresponds to Altaic speakers. It will be extremely interesting to see what the analysis including other Altaic speakers from my other datasets as well as West Eurasians of Uralic/Altaic language or with "Northeast Asian" admixture will show.
The table below has sample sizes and admixture proportions.
Stay tuned. More to come.
October 17, 2010
ADMIXTURE across Eurasia: from Anatolia to Siberia
(Last Update: Oct 17)
Here is a result of an ADMIXTURE run of a few populations from Eurasia (left to right: Turks, Armenians, Georgians, followed by a mix of Uygur, Mongolians, Yakut, Hezhen in no order), combining the HGDP dataset with that of Behar et al. (2010).

It's more of a test, rather than a final result, as I've just finished integrating the two datasets, but it's a nice comparison of a wide assortment of linguistic families.
Notice Turks and Armenians being quite similar to each other, (green+blue), although Turks are differentiated by the presence of an east Eurasian component (5.5%). On the basis of uniparental markers, five years ago, I estimated this component as 6.2% which seems to be right on the money. In the combined Armenian/Georgian sample this admixture is only 0.14% and as can be seen is limited to a handful of Georgian individuals.
It is interesting that Georgians belong semi-uniquely to the green cluster. Turks' non-Mongoloid ancestors were Indo-European speaking like the Armenians still are. It would be tempting to see in the blue-green contrast an Indo-European/Caucasian one, especially as the Caucasoid component further east seems to be mainly blue, in agreement with the idea that it was Indo-Europeans (in particular mainly Iranic speakers) who brought Caucasoid genes to the heartland of Asia.
UPDATE I (Oct 17):
Moving to the north, we see (left-to-right) Han (red), Hungarian/Belorussian (blue), Chuvash (first red "step"), Uzbek (second red "step"). Unlike the Turks, the Hungarians, who also speak a language that came from the east, seem to lack a noticeable east Eurasian component.
Their linguistic conversion was one of elite dominance, where a handful of Mongoloid and quasi-Mongoloid upper echelons left their language but not their genes:
According to his observations, the “overlords” were characterized by Turanid, Uralian and Pamir race elements and also by certain long-headed components. The “middle layer” or “warriors’ layer”, however, showed an anthropological profile distinctly different from that of the overlords. It was essentially constituted by Mediterraneans, Nordoids (who might also have been tall robust Mediterraneans) and Pamir component while the absence of Turanid and Uralian race characteristics was remarkable. As regards the third layer, the so-called “common folk”, they were dominated, just as the middle layer was, by Mediterranean and Nordoid elements but, in addition, the Cromagnoid ones were also significant.
September 30, 2010
More ADMIXTURE estimates in Eurasia
This time, I removed SNPs with more than 1% genotyping no-call from the Xing et al. (2010) dataset, and ran ADMIXTURE on the following populations (left-to-right): Slovenian, Kyrgyzstani, Buryat, and HapMap Chinese. For K=2:
The results are as expected, with Slovenians and Chinese forming opposite poles, and Kyrgyzstanis and Buryat showing a preponderence of Mongoloid ancestry, but with variable Caucasoid admixture. Notice a single Slovenian showing eastern influence.
For K=3:
The Buryat get their own cluster (blue). Some Chinese are seen as having "Buryat" influence, which makes sense as there have been incursion of Mongols into China in historical times. Some Buryat too seem to be "Chinese"-influenced.
Kyrgyzstanis show mixed affiliations. The presence of both a Buryat and a "Chinese" cluster is interesting. The Kyrgyz live at a lower latitude than the Buryat, so this may be a reason behind the "Chinese" cluster, while the Buryat are a more purely northern Mongoloid population.
Notice too, how the lone Slovenian becomes "blue" indicating Mongol rather than Chinese origins. This also makes sense as the Chinese people did not migrate to Europe, while Mongoloids of the steppe and forest zones did.
Interesting is also the emergence of a 2-3 Buryat with some "European" admixture. These may not stem from the centuries old mix between Sakas and Mongols, but may represent a more recent (e.g., Slavic) European element.
September 08, 2008
mtDNA of Mongolians
While I agree that the Mongol Empire played a major role in the admixture of Caucasoids and Mongoloids, we have very clear evidence now that the two races had been mixing in Central Asia long before that time.
J Hum Genet.
Genetic imprint of the Mongol: signal from phylogeographic analysis of mitochondrial DNA
Cheng B, Tang W, He L, Dong Y, Lu J, Lei Y, Yu H, Zhang J, Xiao C.
Abstract
Mitochondrial deoxyribonucleic acid (DNA) from 201 unrelated Mongolian individuals in the three different regions was analyzed. The Mongolians took the dominant East Asian-specific haplogroups, and some European-prevalent haplogroups were detected. The East Asians-specific haplogroups distributed from east to west in decreasing frequencies, and the European-specific haplogroups distributed conversely. These genetic data suggest that the Mongolian empire played an important role in the maternal genetic admixture across Mongolians and even Central Asian populations, whereas the Silk Road might have contributed little in the admixture between the East Asians and the Europeans.
Link
J Hum Genet.
Genetic imprint of the Mongol: signal from phylogeographic analysis of mitochondrial DNA
Cheng B, Tang W, He L, Dong Y, Lu J, Lei Y, Yu H, Zhang J, Xiao C.
Abstract
Mitochondrial deoxyribonucleic acid (DNA) from 201 unrelated Mongolian individuals in the three different regions was analyzed. The Mongolians took the dominant East Asian-specific haplogroups, and some European-prevalent haplogroups were detected. The East Asians-specific haplogroups distributed from east to west in decreasing frequencies, and the European-specific haplogroups distributed conversely. These genetic data suggest that the Mongolian empire played an important role in the maternal genetic admixture across Mongolians and even Central Asian populations, whereas the Silk Road might have contributed little in the admixture between the East Asians and the Europeans.
Link
July 28, 2008
Ancient mtDNA from Inner Mongolia
Three individuals with mixed Caucasoid-Mongoloid affinities were an adult female (haplogroup C), 25yo male (haplogroup M), and 25-30yo male (haplogroup A). From the paper:
Ancient DNA analysis of human remains from the upper capital city of Kublai Khan
Yuqin Fu et al.
Abstract
Analysis of DNA from human archaeological remains is a powerful tool for reconstructing ancient events in human history. To help understand the origin of the inhabitants of Kublai Khan's Upper Capital in Inner Mongolia, we analyzed mitochondrial DNA (mtDNA) polymorphisms in 21 ancient individuals buried in the Zhenzishan cemetery of the Upper Capital. MtDNA coding and noncoding region polymorphisms identified in the ancient individuals were characteristic of the Asian mtDNA haplogroups A, B, N9a, C, D, Z, M7b, and M. Phylogenetic analysis of the ancient mtDNA sequences, and comparison with extant reference populations, revealed that the maternal lineages of the population buried in the Zhenzishan cemetery are of Asian origin and typical of present-day Han Chinese, despite the presence of typical European morphological features in several of the skeletons.
Link
All haplogroups were Asian-specific, the haplotypes of 10 individuals are shared by modern Han Chinese, and the one-step neighbors to another 7 individuals also mainly distribute in modern Han Chinese (Yao et al., 2002). The phylogenetic analysis of the ancient population and extant Eurasian populations showed that the ancient population most closely related to the Han Chinese, especially the northern Han.American Journal of Physical Anthropology doi: 10.1002/ajpa.20894
Ancient DNA analysis of human remains from the upper capital city of Kublai Khan
Yuqin Fu et al.
Abstract
Analysis of DNA from human archaeological remains is a powerful tool for reconstructing ancient events in human history. To help understand the origin of the inhabitants of Kublai Khan's Upper Capital in Inner Mongolia, we analyzed mitochondrial DNA (mtDNA) polymorphisms in 21 ancient individuals buried in the Zhenzishan cemetery of the Upper Capital. MtDNA coding and noncoding region polymorphisms identified in the ancient individuals were characteristic of the Asian mtDNA haplogroups A, B, N9a, C, D, Z, M7b, and M. Phylogenetic analysis of the ancient mtDNA sequences, and comparison with extant reference populations, revealed that the maternal lineages of the population buried in the Zhenzishan cemetery are of Asian origin and typical of present-day Han Chinese, despite the presence of typical European morphological features in several of the skeletons.
Link
July 21, 2008
Y chromosomes and mtDNA of Daghestan groups
This is a free paper which establishes the difference between highland Northeast Caucasian speakers and lowland Altaic speakers in Daghestan. The lowland groups show evidence of Mongoloid haplogroups in both Y chromosomes and mtDNA, while the highland groups are dominated by haplogroup J:
Interestingly, haplogroup G occurs in the Avars (0.06) but not in the other highland groups. Haplogroup G is common in the Southern Caucasus. The mountain groups also have little R1*(xR1a1) (0.06 in Avars, 0.08 in Kubachi) and no I, R1a1 or E.
It certainly seems to be the case that the highland Northeast Caucasian speakers are descended from a J1-dominated ancient Near Eastern population which was preserved due to patrilocal endogamy. The relationship -that I wrote about earlier- of these Caucasian J1's to the Arabian J1's, the second major region of J1 dominance remains to be seen.
BMC Genetics 2008, 9:47 doi:10.1186/1471-2156-9-47
Culture creates genetic structure in the Caucasus: Autosomal, mitochondrial, and Y-chromosomal variation in Daghestan
Elizabeth E Marchani 1, W Scott Watkins 2, Kazima Bulayeva 3, Henry C
Harpending 1, Lynn B Jorde 2§
Abstract
Background
Near the junction of three major continents, the Caucasus region has been an important thoroughfare for human migration. While the Caucasus Mountains have diverted human traffic to the few lowland regions that provide a gateway from north to south between the Caspian and Black Seas, highland populations have been isolated by their remote geographic location and their practice of patrilocal endogamy. We investigate how these cultural and historical differences between highland and lowland populations have affected patterns of genetic diversity. We test 1) whether the highland practice of patrilocal endogamy has generated sex-specific population relationships, and 2) whether the history of migration and military conquest associated with the lowland populations has left Central Asian genes in the Caucasus, by comparing genetic diversity and pairwise population relationships between Daghestani populations and reference populations throughout Europe and Asia for autosomal, mitochondrial, and Y-chromosomal markers.
Results
We found that the highland Daghestani populations had contrasting histories for the mitochondrial DNA and Y-chromosome data sets. Y-chromosomal haplogroup diversity was reduced among highland Daghestani populations when compared to other populations and to highland Daghestani mitochondrial DNA haplogroup diversity. Lowland Daghestani populations showed Turkish and Central Asian affinities for both mitochondrial and Y-chromosomal data sets. Autosomal population histories are strongly correlated to the pattern observed for the mitochondrial DNA data set, while the correlation between the mitochondrial DNA and Y-chromosome distance matrices was weak and not significant.
Conclusions
The reduced Y-chromosomal diversity exhibited by highland Daghestani populations is consistent with genetic drift caused by patrilocal endogamy. Mitochondrial and Ychromosomal phylogeographic comparisons indicate a common Near Eastern origin of highland populations. Lowland Daghestani populations show varying influence from Near Eastern and Central Asian populations.
Link (pdf)
The highland Avar, Dargin, and Kubachi exhibit high frequencies of haplogroup J (0.56, 1.00, and 0.67, respectively)According to Table 2, the Avars possess 0.33 of J2, so, consistent with previous observations, the Northeast Caucasian groups are J1 (or at least J*(xJ2)) exclusive.
Interestingly, haplogroup G occurs in the Avars (0.06) but not in the other highland groups. Haplogroup G is common in the Southern Caucasus. The mountain groups also have little R1*(xR1a1) (0.06 in Avars, 0.08 in Kubachi) and no I, R1a1 or E.
It certainly seems to be the case that the highland Northeast Caucasian speakers are descended from a J1-dominated ancient Near Eastern population which was preserved due to patrilocal endogamy. The relationship -that I wrote about earlier- of these Caucasian J1's to the Arabian J1's, the second major region of J1 dominance remains to be seen.
BMC Genetics 2008, 9:47 doi:10.1186/1471-2156-9-47
Culture creates genetic structure in the Caucasus: Autosomal, mitochondrial, and Y-chromosomal variation in Daghestan
Elizabeth E Marchani 1, W Scott Watkins 2, Kazima Bulayeva 3, Henry C
Harpending 1, Lynn B Jorde 2§
Abstract
Background
Near the junction of three major continents, the Caucasus region has been an important thoroughfare for human migration. While the Caucasus Mountains have diverted human traffic to the few lowland regions that provide a gateway from north to south between the Caspian and Black Seas, highland populations have been isolated by their remote geographic location and their practice of patrilocal endogamy. We investigate how these cultural and historical differences between highland and lowland populations have affected patterns of genetic diversity. We test 1) whether the highland practice of patrilocal endogamy has generated sex-specific population relationships, and 2) whether the history of migration and military conquest associated with the lowland populations has left Central Asian genes in the Caucasus, by comparing genetic diversity and pairwise population relationships between Daghestani populations and reference populations throughout Europe and Asia for autosomal, mitochondrial, and Y-chromosomal markers.
Results
We found that the highland Daghestani populations had contrasting histories for the mitochondrial DNA and Y-chromosome data sets. Y-chromosomal haplogroup diversity was reduced among highland Daghestani populations when compared to other populations and to highland Daghestani mitochondrial DNA haplogroup diversity. Lowland Daghestani populations showed Turkish and Central Asian affinities for both mitochondrial and Y-chromosomal data sets. Autosomal population histories are strongly correlated to the pattern observed for the mitochondrial DNA data set, while the correlation between the mitochondrial DNA and Y-chromosome distance matrices was weak and not significant.
Conclusions
The reduced Y-chromosomal diversity exhibited by highland Daghestani populations is consistent with genetic drift caused by patrilocal endogamy. Mitochondrial and Ychromosomal phylogeographic comparisons indicate a common Near Eastern origin of highland populations. Lowland Daghestani populations show varying influence from Near Eastern and Central Asian populations.
Link (pdf)
March 04, 2008
AAPA 2008 abstracts
The 2008 meeting of the American Association of Physical Anthropologists will take place this April, and the book of abstracts for the conference is online in pdf format. As usual, there is a great variety of exciting research to be announced in the meeting; here is my sampling thereof:
A seemingly very important new piece of work on Central Anatolia:
O. Gokcumen et al., The Land of the Tired Ox: Ethnogenetic Insights into Rural Central Anatolian Population History
I can only hope that more researchers will look into historical processes that have shaped modern populations. Too often I see research published which tries to infer human prehistory from modern populations, seemingly oblivious to the complex set of events in historical time that have shaped these populations. Anatolia, so often discussed in the context of the origin of the Neolithic is a prime example of this, as it contains multiple layers of population settlement and ethnic change.
M. C. Dulik et al. Y-chromosome variation in Altaian ethnic groups
L. Pipes et al. Analysis of mtDNA in Mongolian Populations
J. Hawks. "Adaptive evolution of human hearing and the appearance of language"
B.E. Hemphill. Are the inhabitants of Madaklasht an emigrant Persian population in northern Pakistan?: a dental morphometric investigation.
Someone should look at their genes. Human history is a giant jigsaw puzzle and it is populations that differ from their neighbors and came from somewhere else that allow us to catch a glimpse of the past (in this case prehistoric Central Asia).
N. Seguchi. "Re-analysis of the ainu-samurai hypothesis using population genetic analysis."
A seemingly very important new piece of work on Central Anatolia:
O. Gokcumen et al., The Land of the Tired Ox: Ethnogenetic Insights into Rural Central Anatolian Population History
Excerpt: "For example, in one study area in the vicinity of Ankara, we have observed at least four distinct groups based on historical and ethnographic observations. Their self-claimed ancestries trace back to Afsar, Kurdish, Caucasian Cherkess, and Karaman groups. These groups came into the same area from different source regions and at different moments in history. Indeed, our data indicate that there were significant disparities between the paternal and maternal genetic diversity among these groups. These data also allow us to more accurately reconstruct the population history of the study area, as well as begin to provide new perspectives on the regional history of Central Anatolia in relation to historical Turkic invasions and perhaps the Neolithic transition. Finally, we discuss the utility of a more focal and detailed sampling approach for elucidating Anatolian population history."
I can only hope that more researchers will look into historical processes that have shaped modern populations. Too often I see research published which tries to infer human prehistory from modern populations, seemingly oblivious to the complex set of events in historical time that have shaped these populations. Anatolia, so often discussed in the context of the origin of the Neolithic is a prime example of this, as it contains multiple layers of population settlement and ethnic change.
M. C. Dulik et al. Y-chromosome variation in Altaian ethnic groups
Excerpt: "A large portion of all Altaian haplotypes belonged to haplogroup R. Differences in haplogroup frequency between the northern and southern Altaian populations were also observed, with more individuals from northern groups belonging to haplogroups N and Q, and haplogroup C being more prevalent in southern populations. In addition, there were village level patterns of NRY variation, while the overall diversity of NRY haplotypes suggested a significant cultural influence on the partitioning of genetic variation (i.e., patrilocality)."The three elements involved in Siberian prehistory are indeed haplogroup R, in particular R1a1 which (in my opinion) represents the Western-derived Caucasoid element of likely Iranic affiliation, haplogroups N and Q which represent the Palaeo-Mongoloid element indigenous to Siberia and which has radiated from Siberia to the west (in the case of N) and to the east and into the Americas (in the case of Q), and the Mongoloid proper element which is associated with haplogroup C in this region, and which reflects the Eastern-derived movements of Mongoloid(-influenced) Altaic speakers such as the Mongols.
L. Pipes et al. Analysis of mtDNA in Mongolian Populations
J. Hawks. "Adaptive evolution of human hearing and the appearance of language"
Language requires not only a detailed anatomical and neurological system of language production, but also a highly adapted system of reception. Considering the frequency and amplitude range of human speech, the necessity of perceiving a large number of distinct speakers, the extended life history of humans, the need for children to learn phonemic distinctions at an early age, and the spatial distances covered by vocal communication in humans compared to other primates, it is likely that humans have distinctive auditory adaptations to language. This study tests the hypothesis of selection on the human auditory system, by interspecific genomic comparisons and genome-wide selection scans in living people. A set of hearing-related human genes shows clear signs of recurrent selected substitutions in humans compared to chimpanzees and macaques. These recurrent substitutions may have occurred at any time during human evolutionary history, but they were repeated with several selected variants for each gene. A smaller set of genes shows signs of significant population differentiation within the past 50,000 years, due to recent strong selection. Further, a relatively large set of hearing-related genes have segregating variants under recent strong selection in one or more human populations. These genes reflect continuing selection on hearing within the last 2000—3000 years. Together, these results suggest that human vocal communication exerted repeated selection pressures on the auditory system, that the system of human language continued to evolve during the Late Pleistocene, and that humans may still be adapting to language.It seems that Hawks et al. paper on accelerated recent human evolution was just the beginning...
B.E. Hemphill. Are the inhabitants of Madaklasht an emigrant Persian population in northern Pakistan?: a dental morphometric investigation.
The answer: "Madaklasters share closest affinities to prehistoric Central Asians and more distant affinities to prehistoric inhabitants of the Iranian Plateau. Such results support the claim that the inhabitants of Madaklast are an intrusive population into Pakistan whose origins most likely may be found in northeastern Afghanistan and Tajikistan."
Someone should look at their genes. Human history is a giant jigsaw puzzle and it is populations that differ from their neighbors and came from somewhere else that allow us to catch a glimpse of the past (in this case prehistoric Central Asia).
N. Seguchi. "Re-analysis of the ainu-samurai hypothesis using population genetic analysis."
The conclusion: "The result shows that the Kamakura ties to the Ainu first, before it ties to the other ethnic Japanese. In addition, the Kamakura group shows more variability,indicating that the Kamakura group may have experienced significantly more gene flow. This indicates the Ainu-derived people who lived in East Japan at that time made a genetic contribution to the warrior class of Kamakura."J. K. Rilling et al. "Abdominal depth as a principal determinant of human female attractiveness."
Excerpt: "Multiple linear regression analysis revealed that the depth of the lower torso at the umbilicus, or abdominal depth, was the strongest predictor of attractiveness, stronger than either BMI or WHR, and that its impact was significantly greater for video and side view stimuli in which it was clearly visible compared with front and back view stimuli. Women with shallow abdominal depth are more likely to be healthy, fertile and non-pregnant, suggesting that this may be an adaptive male preference that has been shaped by natural selection."
May 21, 2007
Distribution of Genghis Khan's descendants
Genetika. 2007 Mar;43(3):422-6.
[Distribution of the male lineages of Genghis Khan's descendants in northern Eurasian populations]
[Article in Russian]
[No authors listed]
Data on the variation of 12 microsatellite loci of Y-chromosome haplogroup C3 were used to screen lineages included in the cluster of Genghis Khan's descendants in 18 northern Eurasian populations (Altaian Kazakhs, Altaians-Kizhi, Teleuts, Khakassians, Shorians, Tyvans, Todjins, Tofalars, Sojots, Buryats, Khamnigans, Evenks, Mongols, Kalmyks, Tajiks, Kurds, Persians, and Russians; the total sample size was 1437 people). The highest frequency of haplotypes from the cluster of the Genghis Khan's descendants was found in Mongols (34.8%). In Russia, this cluster was found in Altaian Kazakhs (8.3%), Altaians (3.4%), Buryats (2.3%), Tyvans (1.9%), and Kalmyks (1.7%).
Link
[Distribution of the male lineages of Genghis Khan's descendants in northern Eurasian populations]
[Article in Russian]
[No authors listed]
Data on the variation of 12 microsatellite loci of Y-chromosome haplogroup C3 were used to screen lineages included in the cluster of Genghis Khan's descendants in 18 northern Eurasian populations (Altaian Kazakhs, Altaians-Kizhi, Teleuts, Khakassians, Shorians, Tyvans, Todjins, Tofalars, Sojots, Buryats, Khamnigans, Evenks, Mongols, Kalmyks, Tajiks, Kurds, Persians, and Russians; the total sample size was 1437 people). The highest frequency of haplotypes from the cluster of the Genghis Khan's descendants was found in Mongols (34.8%). In Russia, this cluster was found in Altaian Kazakhs (8.3%), Altaians (3.4%), Buryats (2.3%), Tyvans (1.9%), and Kalmyks (1.7%).
Link
Subscribe to:
Posts (Atom)