April 24, 2016
Jewish and Indian ancestry in the Bene Israel
The Genetics of Bene Israel from India Reveals Both Substantial Jewish and Indian Ancestry
Yedael Y. Waldman , Arjun Biddanda , Natalie R. Davidson, Paul Billing-Ross, Maya Dubrovsky, Christopher L. Campbell, Carole Oddoux, Eitan Friedman, Gil Atzmon, Eran Halperin, Harry Ostrer, Alon Keinan
The Bene Israel Jewish community from West India is a unique population whose history before the 18th century remains largely unknown. Bene Israel members consider themselves as descendants of Jews, yet the identity of Jewish ancestors and their arrival time to India are unknown, with speculations on arrival time varying between the 8th century BCE and the 6th century CE. Here, we characterize the genetic history of Bene Israel by collecting and genotyping 18 Bene Israel individuals. Combining with 486 individuals from 41 other Jewish, Indian and Pakistani populations, and additional individuals from worldwide populations, we conducted comprehensive genome-wide analyses based on FST, principal component analysis, ADMIXTURE, identity-by-descent sharing, admixture linkage disequilibrium decay, haplotype sharing and allele sharing autocorrelation decay, as well as contrasted patterns between the X chromosome and the autosomes. The genetics of Bene Israel individuals resemble local Indian populations, while at the same time constituting a clearly separated and unique population in India. They are unique among Indian and Pakistani populations we analyzed in sharing considerable genetic ancestry with other Jewish populations. Putting together the results from all analyses point to Bene Israel being an admixed population with both Jewish and Indian ancestry, with the genetic contribution of each of these ancestral populations being substantial. The admixture took place in the last millennium, about 19–33 generations ago. It involved Middle-Eastern Jews and was sex-biased, with more male Jewish and local female contribution. It was followed by a population bottleneck and high endogamy, which can lead to increased prevalence of recessive diseases in this population. This study provides an example of how genetic analysis advances our knowledge of human history in cases where other disciplines lack the relevant data to do so.
Link
December 18, 2013
Near Eastern origin of R1a in Ashkenazi Levites
It turns out that this is not the case. The specific clade R-M582 to which Ashkenazi Levites (and other non-Levites) belong to is absent in eastern Europeans and present in non-Jewish Near Easterners, making it more likely that Jews did not pick it up from eastern Europeans, but rather from some Near Eastern population. A look at the table of frequencies suggests to me an Iranic source, but I doubt that modern populations will ever allow a full resolution of such questions.
Nature Communications 4, Article number: 2928 doi:10.1038/ncomms3928
Phylogenetic applications of whole Y-chromosome sequences and the Near Eastern origin of Ashkenazi Levites
Siiri Rootsi et al.
Previous Y-chromosome studies have demonstrated that Ashkenazi Levites, members of a paternally inherited Jewish priestly caste, display a distinctive founder event within R1a, the most prevalent Y-chromosome haplogroup in Eastern Europe. Here we report the analysis of 16 whole R1 sequences and show that a set of 19 unique nucleotide substitutions defines the Ashkenazi R1a lineage. While our survey of one of these, M582, in 2,834 R1a samples reveals its absence in 922 Eastern Europeans, we show it is present in all sampled R1a Ashkenazi Levites, as well as in 33.8% of other R1a Ashkenazi Jewish males and 5.9% of 303 R1a Near Eastern males, where it shows considerably higher diversity. Moreover, the M582 lineage also occurs at low frequencies in non-Ashkenazi Jewish populations. In contrast to the previously suggested Eastern European origin for Ashkenazi Levites, the current data are indicative of a geographic source of the Levite founder lineage in the Near East and its likely presence among pre-Diaspora Hebrews.
Link
October 08, 2013
Ashkenazi Jewish matrilineages mainly of European origin
If we allow for the possibility that K1a9 and N1b2 might have a Near Eastern source, then we can estimate the overall fraction of European maternal ancestry at ~65%. Given the strength of the case for even these founders having a European source, however, our best estimate is to assign ~81% of Ashkenazi lineages to a European source, ~8% to the Near East and ~1% further to the east in Asia, with ~10% remaining ambiguous (Fig. 10; Supplementary Table S9). Thus at least two-thirds and most likely more than four-fifths of Ashkenazi maternal lineages have a European ancestry.Nature Communications 4, Article number: 2543 doi:10.1038/ncomms3543
A substantial prehistoric European ancestry amongst Ashkenazi maternal lineages
Marta D. Costa et al.
The origins of Ashkenazi Jews remain highly controversial. Like Judaism, mitochondrial DNA is passed along the maternal line. Its variation in the Ashkenazim is highly distinctive, with four major and numerous minor founders. However, due to their rarity in the general population, these founders have been difficult to trace to a source. Here we show that all four major founders, ~40% of Ashkenazi mtDNA variation, have ancestry in prehistoric Europe, rather than the Near East or Caucasus. Furthermore, most of the remaining minor founders share a similar deep European ancestry. Thus the great majority of Ashkenazi maternal lineages were not brought from the Levant, as commonly supposed, nor recruited in the Caucasus, as sometimes suggested, but assimilated within Europe. These results point to a significant role for the conversion of women in the formation of Ashkenazi communities, and provide the foundation for a detailed reconstruction of Ashkenazi genealogical history.
Link
November 22, 2012
ALDER signal of admixture in Ashkenazi Jews
Previous studies on uniparental markers have indicated that Ashkenazi Jews (AJ) were formed by admixture between a Near Eastern population and European host populations; the evidence for the former element seems pretty clear on the basis of Y-chromosomes where Jews possess a relatively high frequency of Y-haplogroup J1 (and a few others) that are quite rare in non-Jewish north/east Europeans. As for the latter, it seems probable on the basis of the location of Ashkenazi Jews on PCA plots where they tend to occupy an intermediate position between extant populations of the Levant (including Near Eastern Jews) and non-Jewish Europeans.
Anyone who has played around with genetic data will know that while AJ may be positioned in the aforementioned "intermediate" location within the "West Eurasian continuum" between Europe and Near East, they tend to form their own cluster at higher dimensions. And, indeed, this is why it's fairly easy for a clustering algorithm, such as my "Clusters Galore" (MCLUST/MDS) approach to pick out a very specific AJ cluster (e.g., here, or here, using a fastIBD approach). An Ashkenazi Jewish-specific cluster also pops out at higher K in ADMIXTURE analyses. This cluster may reflect endogamy within the AJ community until quite recent times.
One way of detecting admixture in a group is through the use of f3-statistics. The statistic f3(AJ; European, Near_East) could be negative --which would indicate admixture-- but it is usually not -at least in the combinations of (European, Near_East) I've tried, and this is consistent with either the presence admixture or absence of admixture.
A simple and intuitive way to see why post-admixture drift might mask the presence of admixture can be seen by means of a simple calculation. Remember that the f3-statistic's +/- sign depends on the +/- sign of quantities (c-a)*(c-b) where c is an allele frequency in the admixed (?) population we are investigating, and a, b in the two reference populations. We can pick a to be less than b with no loss of generality.
In the absence of strong drift (e.g., if all populations have a very large number of individuals), then the allele frequency c=xa+(1-x)b where x is the amount of admixture --between 0 and 1-- from group A and (1-x) from group B, and this c will be maintained little changed in the post-admixture phase. With the aid of a little algebra, we get that:
(c-a)*(c-b) = (xa+(1-x)b-a)*(xa+(1-x)b-b)
= (xa+b-xb-a)*(xa+b-xb-b) =
= x(x-1)(a-b)^2
and this is of course negative because we assumed that x was less than 1.
In a large population, this c will remain near-constant, because of the lack of strong drift. As long as it remains within the interval (a,b), then (c-a)*(c-b) will also remain negative, and so will the f3 statistic.
But, what if strong drift affects the admixed population? Allele frequencies fluctuate more wildly in larger populations, so c might go outside the (a,b) interval. Without loss of generality, assume that c becomes greater than b in which case (c-a)*(c-b) will become positive.
The f3-statistic averages over many SNPs, so, depending on (i) the initial differentiation of the admixed populations, which could be seen as b-a, and (ii) the amount of drift, which causes c to jump outside the (a, b) interval as discussed above, it is possible that the evidence for admixture may disappear.
So, relying on allele frequency differences may help obliterate the signal of admixture. But, there is a different signal of admixture that uses the decay of admixture linkage-disequilibrium, most recently discussed in the ALDER paper. The admixture LD signal's evidence may also disappear in time, but only because the signal occurs at increasingly lower genetic distances over time due to recombination. Thankfully, it tends to occur at large enough --for the last few thousand years-- distances, for which the SNP density of existing genotyping platforms that measure a few hundred thousand SNPs per individual is sufficient.
METHODS
Naturally I was curious to see whether the admixture LD mechanism would produce the evidence of admixture that the f3-statistics did not. I combined three datasets in my possession (HGDP by Li et al. Behar et al. and Yunusbayev et al. ) and identified sets of European and Semitic populations. (Remember that these sets are non-exhaustive, but presumably usable surrogates for the true mixing populations exist within them):
Abhkasians_Y, Adygei, Belorussian, Bulgarians_Y, Chechens_Y, Chuvashs, French, French_Basque, Georgians, Hungarians, Lezgins, Lithuanians, Mordovians_Y, North_Italian, North_Ossetians_Y, Orcadian, Romanians, Russian, Sardinian, Spaniards, Tuscan, Ukranians_Y
and:
Bedouin, Druze, Egyptans, Ethiopian_Jews, Ethiopians, Iraq_Jews, Jordanians, Lebanese, Morocco_Jews, Palestinian, Saudis, Sephardic_Jews, Syrians, Yemenese, Yemen_Jews
I used my Dodecad Project sample of AJ which numbers 36 individuals and is larger than any other usable public sample available to me.
(ALDER was run with default parameters, using the Rutgets recombination map for Illumina chips, and with the merged dataset prepared with a --geno 0.03 flag. Note that the Ashkenazi_D sample consists of individuals typed on different Illumina platforms from 23andMe and FamilyTreeDNA. The total number of SNPs considered was 527,165.)
RESULTS
I report below the tests for which ALDER reported "success" for the test with no warnings:
The median of all these estimates is 36.78 generations or 1070 years which corresponds to a calendar date of 910CE, assuming the sample's birthday was 1980, and a generation length of 29 years.
Palamara et al. placed the beginning of demographic expansion of AJ in a similar timeframe (33 generations), following a severe founder effect reducing the population to ~270 individuals. Such a founder effect may have indeed served to produce positive f3-statistics, masking the presence of admixture, the occurrence of which appears to be substantiated on the basis of the ALDER test of admixture.
As for the levels of admixture, using a 1-ref analysis with the European populations, I get the following lower bounds:
I'd be interested in hearing people's opinions on the plausibility of these dates/proportions, as well as their potential historical associations; a lot of factors might affect these results, so perhaps this analysis could be improved in the future.
October 26, 2012
IBD length distribution and demographic history (Palamara)
In any case, the conclusions are worth mentioning in themselves. For the Ashkenazi:
We obtained an improved fit for a population composed of ~2,300 ancestors 200 generations before the present; this population exponentially expanded to reach ~45,000 individuals 34 generations ago. After a severe founder event, the population was reduced to ~270 individuals, which then expanded rapidly during 33 generations (rate r ~ 0.29) and reached a modern population of ~4,300,000 individuals.And, for the Maasai:
Optimizing a model of exponential expansion and contraction (Figure 1A), we obtained a good fit to the observed IBD frequency spectrum (Figure 6), suggesting that an ancestral population of ~23,500 individuals decreased to ~500 current individuals during the course of 23 generations (r ~ -0.17). We note that this result might not be driven by an actual gradual population contraction in the MKK individuals, but it most likely reflects the societal structure of this seminomadic population. ... We thus used the village model to analyze the MKK demography and relied on coalescent simulations to retrieve its parameters: migration rate, size, and number of villages that provide a good fit for the empirical distribution of IBD segments.We observed a compatible fit for this model, in which 44 villages of 485 individuals each intermix with a migration rate of 0.13 individuals per generation (Figure 6).If I understand this correctly, it appears that Maasai (MKK) individuals share long IBD segments not because their population has contracted (and hence they're all descended from a limited number of founders, as is the case for Ashkenazi Jews), but rather because their social structure follows the "village model" in which people share shallow ancestry (and hence long IBD) with other people in their "village" and exchange genes with other "villages".
The American Journal of Human Genetics, 25 October 2012 doi:10.1016/j.ajhg.2012.08.030
Length Distributions of Identity by Descent Reveal Fine-Scale Demographic History
Pier Francesco Palamara et al.
Data-driven studies of identity by descent (IBD) were recently enabled by high-resolution genomic data from large cohorts and scalable algorithms for IBD detection. Yet, haplotype sharing currently represents an underutilized source of information for population-genetics research. We present analytical results on the relationship between haplotype sharing across purportedly unrelated individuals and a population’s demographic history. We express the distribution of IBD sharing across pairs of individuals for segments of arbitrary length as a function of the population’s demography, and we derive an inference procedure to reconstruct such demographic history. The accuracy of the proposed reconstruction methodology was extensively tested on simulated data. We applied this methodology to two densely typed data sets: 500 Ashkenazi Jewish (AJ) individuals and 56 Kenyan Maasai (MKK) individuals (HapMap 3 data set). Reconstructing the demographic history of the AJ cohort, we recovered two subsequent population expansions, separated by a severe founder event, consistent with previous analysis of lower-throughput genetic data and historical accounts of AJ history. In the MKK cohort, high levels of cryptic relatedness were detected. The spectrum of IBD sharing is consistent with a demographic model in which several small-sized demes intermix through high migration rates and result in enrichment of shared long-range haplotypes. This scenario of historically structured demographies might explain the unexpected abundance of runs of homozygosity within several populations.
Link
August 08, 2012
fastIBD analysis of several Jewish and non-Jewish groups
- fastIBD was run with default parameters over a dataset of 512 individuals/264,539 SNPs
- fastIBD identifies segments of relatively recent origin that are shared by individuals. These results should not be construed as measures of overall genetic similarity or origins. Rather, they suggest which populations have exchanged genes in the relative recent past, say, the last two thousand years or so.
- I included all Ashkenazi_D and North_African_Jews_D samples; of the other Dodecad and reference populations, I took random samples of 10 each; running time of fastIBD increases with the square of the number of individuals, so doing this allowed me to run this in less than a day as opposed to about a week.
- Spreadsheet of numeric results, showing sharing (in centi-Morgans, cM)
- Population-level graphical results, showing an ordering of other populations based on mean IBD sharing.
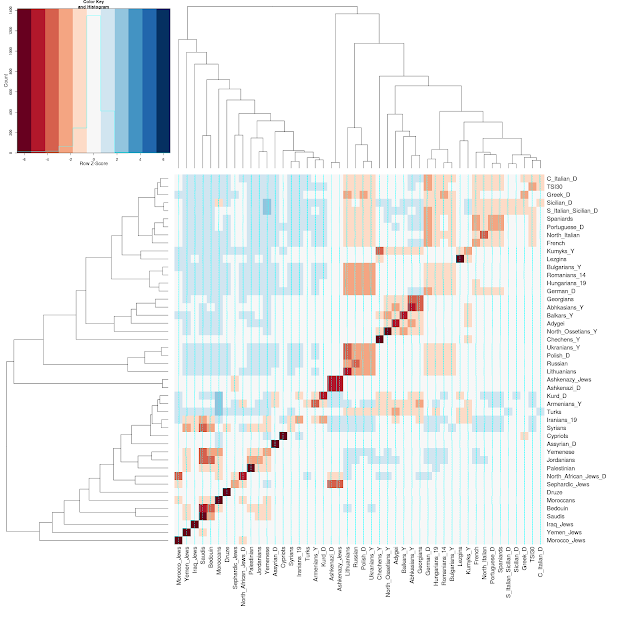
And, here are a couple of the visualizations for a few Jewish populations:
Note that all sources of data are listed on the bottom left of the Dodecad blog.
August 07, 2012
Origins of North African and Central/East European Jews
Some related analyses of mine:
- fastIBD analysis of Afroasiatic groups (Jews, Arabs, Assyrians, Berbers, Somalis, Amharas, etc.)
- fastIBD analysis of Iberia, France, Italy, Balkans, Anatolia and European Jews
- Admixture proportions for various populations (incl. various Jewish groups)
Below is Fig. 3 from Campbell et al.:
One can see that Jewish groups have high degree of intra-population IBD sharing (A); many of the highest levels of IBD sharing is between Jewish groups (B and C).
This paper definitely shows that Jewish groups differ from non-Jewish North Africans. But, the lack of comparative samples from non-Jewish non-North Africans makes the interpretation of this result difficult. Both the PCA analysis, shown below, and the structure analysis indicates a significant Sub-Saharan component in North African non-Jewish populations.
So, it seems, based on these results, that Jewish groups are differentiated from North Africans due to their general lack of sub-Saharan admixture, and they also show a variable degree of affiliation to European groups; however, by "European" groups we go only as far as north Italy and Sardinia. What of the relationships of different Jewish groups to people from southern Italy, Greece, Anatolia, the Caucasus, or even Iranian speakers of the Near East?
Now, let's go to the Elhaik paper, which investigates a different problem altogether, trying to distinguish between the "Rhineland" and "Khazarian" hypotheses for the origins of Central-East European Jews. According to the paper:
Admixture calculations were carried out using a supervised learning approach in a structure-like analysis. This approach has many advantages over the unsupervised approach that not only traces ancestry to K abstract unmixed populations under the assumption that they evolved independently (Chakravarti 2009; Weiss and Long 2009) but also problematic when applied to study Jewish ancestry, which can be dated as far back as 3,000 years (Figure 2). Admixture was calculated with a reference set of seven populations representing genetically distinct regions: Pygmies (Africa), French Basque (West Europe), Chuvash (East Europe), Han Chinese (Asia), Palestinians (Middle East), Turk-Iranians (Near East), and Armenians (Caucasus) (Figure 5).But, Palestinians too have African admixture, so using them as a parental population conflates two separate issues: their old Near Eastern Semitic ancestors which could be reasonably inferred to be somewhat related to the Semitic ancestors of Jews, and their recent African admixture. Similarly, Turks have east Eurasian admixture, and Iranians have South Asian admixture.
The IBD sharing is probably the strongest piece of evidence in this paper for a Caucasian connection. Excess of IBD sharing with Caucasus and Palestinians relative to the other populations may indeed be a good indication of such admixture. On the other hand, the Khazarian Empire was primarily located in eastern Europe and the North Caucasus, not in Armenia and Georgia. Also, this analysis rejects the Greco-Roman hypothesis (whereby European Jews underwent admixture in Greco-Roman times when they were part of the Hellenistic and Roman Empires), but does not really include any Greco-Roman populations (for example, from Greece and Italy).
On the other hand, there may be something to the Khazar story (but in the sense of admixture, rather than replacement). High IBD sharing with Caucasians is one such piece of evidence. Another is the presence of Y-haplogroup Q and R-Z93+, both of which could in principle track a Central Asian Turkic influence (although Z93 could also track an Iranian influence). Then, there is the limited but persistent evidence for a little East Eurasian admixture present in Ashkenazi Jews and not in Sephardic Jews, which might also be consistent with a little Turkic influence.
Overall, I am convinced that most modern Jewish groups have some variable old Near Eastern Jewish ancestry, primarily on the basis of the elevated "Southwest Asian" that seems to correlate reasonably well with groups of Semitic speakers. But, it is difficult to say "how much" and to identify all the potential sources of admixture. Jews have been an international people for quite a long time, so I would guess that fragments of different peoples they encountered may remain in their genomes. Perhaps something akin to Ralph and Coop (2012) may give more information about the timing of these admixture events, as well as the date of the common ancestry of different Jewish groups.
PS: I started a small fastIBD analysis of different Jewish and non-Jewish groups with a fairly large assortment of populations, and will probably post it here in the next few days.
PNAS doi: 10.1073/pnas.1204840109
North African Jewish and non-Jewish populations form distinctive, orthogonal clusters
Christopher L. Campbell et al.
North African Jews constitute the second largest Jewish Diaspora group. However, their relatedness to each other; to European, Middle Eastern, and other Jewish Diaspora groups; and to their former North African non-Jewish neighbors has not been well defined. Here, genome-wide analysis of five North African Jewish groups (Moroccan, Algerian, Tunisian, Djerban, and Libyan) and comparison with other Jewish and non-Jewish groups demonstrated distinctive North African Jewish population clusters with proximity to other Jewish populations and variable degrees of Middle Eastern, European, and North African admixture. Two major subgroups were identified by principal component, neighbor joining tree, and identity-by-descent analysis—Moroccan/Algerian and Djerban/Libyan—that varied in their degree of European admixture. These populations showed a high degree of endogamy and were part of a larger Ashkenazi and Sephardic Jewish group. By principal component analysis, these North African groups were orthogonal to contemporary populations from North and South Morocco, Western Sahara, Tunisia, Libya, and Egypt. Thus, this study is compatible with the history of North African Jews—founding during Classical Antiquity with proselytism of local populations, followed by genetic isolation with the rise of Christianity and then Islam, and admixture following the emigration of Sephardic Jews during the Inquisition.
Link
arXiv:1208.1092v1 [q-bio.PE]
The Missing Link of Jewish European Ancestry: Contrasting the Rhineland and the Khazarian Hypotheses
Eran Elhaik
The question of Jewish ancestry has been the subject of controversy for over two centuries and has yet to be resolved. The "Rhineland Hypothesis" proposes that Eastern European Jews emerged from a small group of German Jews who migrated eastward and expanded rapidly. Alternatively, the "Khazarian Hypothesis" suggests that Eastern European descended from Judean tribes who joined the Khazars, an amalgam of Turkic clans that settled the Caucasus in the early centuries CE and converted to Judaism in the 8th century. The Judaized Empire was continuously reinforced with Mesopotamian and Greco-Roman Jews until the 13th century. Following the collapse of their empire, the Judeo-Khazars fled to Eastern Europe. The rise of European Jewry is therefore explained by the contribution of the Judeo-Khazars. Thus far, however, their contribution has been estimated only empirically; the absence of genome-wide data from Caucasus populations precluded testing the Khazarian Hypothesis. Recent sequencing of modern Caucasus populations prompted us to revisit the Khazarian Hypothesis and compare it with the Rhineland Hypothesis. We applied a wide range of population genetic analyses - including principal component, biogeographical origin, admixture, identity by descent, allele sharing distance, and uniparental analyses - to compare these two hypotheses. Our findings support the Khazarian Hypothesis and portray the European Jewish genome as a mosaic of Caucasus, European, and Semitic ancestries, thereby consolidating previous contradictory reports of Jewish ancestry.
Link
November 28, 2011
Sephardic signature within mtDNA haplogroup T (?)
chondrial DNA."
From the paper:
four avenues are pursued: (1) A search is conducted throughout multiple databases of the first control region of mitochondrial DNA for the T2e5 motif to ascertain the prevalence and geographic affiliation of the new haplotype. (2) One T2e5 sample isWith respect to (1), the author writes:
sequenced for polymorphisms along the entire mitochondrial DNA and compared with T2e sequences to identify any potential coding region mutations that are important for the Sephardic sequence and its relation to other branches. (3) A phylogenetic tree is built from T2e control sequences to provide further information on the relation among lineages including the Sephardic cluster. Although full genomic sequences are usually preferable to avoid misclassifications based on control region information alone, T2e is an ideal subhaplogroup to exploit the more abundant control region data because it is defined by mutations in the control regions alone. Time to the most recent common ancestor is estimated to address questions of when the lineage emerged as well as where. (4) The frequencies of T sub-haplogroups are compared across growing published literature of various populations including from Europe, the Americas, and the Near East. Although the geographic distribution of haplogroup T has been investigated, less is known about the different subhaplogroups, especially T2e.
The combined databases do not appear to have any biases for Iberia, Mexico, or Sephardim.
Stronger evidence for the Sephardic-ness of the haplotype in question could be arrived by dating it to a period consistent with the origins of that population. However:
Time estimates to the most recent common ancestor of the Sephardic signature T2e5 ranged all the way from after the expulsion – clearly impossible – to 415 000 years before present (YBP) (Fast: 338 YBP, 95% confidence interval (95% CI)=present to 763 YBP; Intermediate: 688 YBP, 95% CI=present-3820 YBP; slow: 6811 YBP, CI1=present to 15 245). Given mutations rates that vary by two orders of magnitude,22 as well as other issues with mutation rates and the rho statistic,23,41 at present coalescence analysis cannot be used to distinguish between different plausible timelines for the proposed Sephardic cluster.
The small T2e5 cluster satisfies criteria for being a signature. Although it is premature to set specific thresholds of a signature, a sample of 25% known Sephardic and 50% suspicion of Sephardic origin is overwhelmingly above what would be expected for a general European haplogroup.
These include Jewish settlers seeking asylum after destruction of temples in Jerusalem by Romans and Babylonians 2000–2500 years ago, slightly earlier Jewish settlers in Iberia,7,43 non-Jewish Muslims in the dispersal of Islam 1000+ years ago, non-Jewish Iberian peopling 2500+ years ago that predates all Jewish influx,44 and settlers in Iberia (or Italy) 45000 years ago that entirely predate the existence of Jewish groups. Thus, what is arguably the most contentious issue of whether there is genetic evidence of original Jewish DNA for the Sephardic line cannot be resolved.
Sephardic signature in haplogroup T mitochondrial DNA
Felice L Bedford
Abstract
A rare combination of mutations within mitochondrial DNA subhaplogroup T2e is identified as affiliated with Sephardic Jews, a group that has received relatively little attention. Four investigations were pursued: Search of the motif in 250 000 control region records across 8 databases, comparison of frequencies of T subhaplogroups (T1, T2b, T2c, T2e, T4, T*) across 11 diverse populations, creation of a phylogenic median-joining network from public T2e control region entries, and analysis of one Sephardic mitochondrial full genomic sequence with the motif. It was found that the rare motif belonged only to Sephardic descendents (Turkey, Bulgaria), to inhabitants of North American regions known for secret Spanish–Jewish colonization, or were consistent with Sephardic ancestry. The incidence of subhaplogroup T2e decreased from the Western Arabian Peninsula to Italy to Spain and into Western Europe. The ratio of sister subhaplogroups T2e to T2b was found to vary 40-fold across populations from a low in the British Isles to a high in Saudi Arabia with the ratio in Sephardim more similar to Saudi Arabia, Egypt, and Italy than to hosts Spain and Portugal. Coding region mutations of 2308G and 14499T may locate the Sephardic signature within T2e, but additional samples and reworking of current T2e phylogenetic branch structure is needed. The Sephardic Turkish community has a less pronounced founder effect than some Ashkenazi groups considered singly (eg, Polish), but other comparisons of interest await comparable averaging. Registries of signatures will benefit the study of populations with a large number of smaller-size founders.
Link
April 28, 2011
Comparing five methods of admixture estimation
November 30, 2010
Cluster galore: re-analysis of Behar et al. (2010) data
November 01, 2010
Joe Pickrell redux
Second, Dienekes followed up on his analysis of the ancestry of the GNZ participants with a much larger data set, including individuals of southwest European descent. As expected, when including more data, there was no evidence that Vincent has any Ashkenazi ancestry. Unexpectedly, this was not true for me—even in this larger analysis, the evidence for Ashkenazi ancestry didn’t disappear.
...
As I was mulling over these sorts of issues, I sent the link to my previous analysis to a family member. I didn’t really expect this person to find it that interesting, but hey, you never know. I then got a phone call. I’ll summarize a couple days worth of moderate confusion, second-hand reports of conversations with distant relatives, and family intrigue with this: as it turns out, one of my great-grandparents was indeed a Polish Ashkenazi Jew who immigrated to the United States around the turn of the century. I, obviously, was completely unaware of this.
So to conclude, a tip of my hat to Dienekes and everyone else who looked at these data—this has been the first genuinely unexpected thing to come out of my genetic data.I've estimated Joe's ancestry here and here.
He is included in the Dodecad Project's spreadsheet as JKP001.
October 18, 2010
Joe Pickrell on his ancestry
a European-origin component in the composite Ashkenazi Jewish gene pool that he happens to share.
October 11, 2010
Running EURO-DNA-CALC on GenomesUnzipped
August 26, 2010
Analysis of Ashkenazi Jewish genomes (Bray et al. 2010)
Investigators in the laboratory of Stephen Warren, PhD, chairman of human genetics at Emory University School of Medicine, used DNA microarray technology to read variant sites across the entire genomes of 471 Ashkenazi Jews. The work comes from a collaboration between Warren and Ann Pulver, ScD, associate professor of psychiatry and behavioral sciences at Johns Hopkins University School of Medicine, who recruited the participants for a study of schizophrenia genetics.
Researchers looked for close to one million single nucleotide polymorphisms (SNPs): common alternative spellings in the genome, analogous to American and British spellings of words such as organize/organise. One measure of genetic diversity in a population is heterozygosity, or how many of the SNPs inherited from the mother and father are different; a more inbred population has less heterozygosity.
"We were surprised to find evidence that Ashkenazi Jews have higher heterozygosity than Europeans, contradicting the widely-held presumption that they have been a largely isolated group," says first author Steven Bray, PhD, a postdoctoral fellow in Warren's laboratory.
...
High linkage disequilibrium can come either from an isolated population (for example, an island whose residents are all descendents of shipwreck survivors) or the relatively recent mixture of separate populations. Bray and his colleagues did find evidence of elevated linkage disequilibrium in the Ashkenazi Jewish population, but were able to show that this matches signs of interbreeding or "admixture" between Middle Eastern and European populations.
The researchers were able to estimate that between 35 and 55 percent of the modern Ashkenazi genome comes from European descent.
"Our study represents the largest cohort of Ashkenazi Jews examined to date with such a high density of genetic markers, and our estimate of admixture is considerably higher than previous estimates that used the Y chromosome to calculate European admixture at between five and 23 percent," Bray says.
...
The new paper comes in the heels of two other papers by Behar et al. and Atzmon et al. which considered Jews in general, discovering additional clusters of Jews that were distinct from Ashkenazi Jews. As I have argued in my review of these papers, the different clusters are not the result of isolation, as the different groups of Jews do not only deviate from each other, but also in the direction of their host populations. It would be worthwhile to perform similar admixture analyses on non-AJ populations to determine what their influence from host populations is. With a little effort it would be possible to reconstruct the ancestral Jewish population, by identifying what is common in the different Jewish populations.
"Only six of the 21 disease genes that we examined showed evidence of selection," Bray says. "This supports the argument that most of the Ashkenazi-prevalent diseases are not generally being selected for, but instead are likely a result of a genetic bottleneck effect, followed by random drift."

From the paper:
The fixation index, FST, calculated concurrently to the PCA, confirms that there is a closer relationship between the AJ and several European populations (Tuscans, Italians, and French) than between the AJ and Middle Eastern populations (Fig. S2B). This finding can be visualized with a phylogenetic tree built using the FST data (Fig. S2C), showing that the AJ population branches with the Europeans and not Middle Easterners. Two recent studies performing PCA and population clustering with high-density SNP genotyping from many Jewish Diaspora populations, both showed that of the Jewish populations, the Ashkenazi consistently cluster closest to Europeans (13, 25). Genetic distances calculated by both groups also show that the Ashkenazi are more closely related to some host Europeans than to the ancestral Levant (13, 25). Although the proximity of the AJ and Italian populations could be explained by their admixture prior to the Ashkenazi settlement in Central Europe (13), it should be noted that different demographic models may potentially yield similar principal component projections (33); thus, it is also consistent that the projection of the AJ populations is primarily the outcome of admixture with Central and Eastern European hosts that coincidentally shift them closer to Italians along principle component axes relative to Middle Easterners. Taken as a whole, our results, along with those from previous studies, support the model of a Middle Eastern origin of the AJ population followed by subsequent admixture with host Europeans or populations more similar to Europeans. Our data further imply that modern Ashkenazi Jews are perhaps even more similar with Europeans than Middle Easterners.The bolded part reminds me of what I wrote in my review of Atzmon et al. regarding the choice of parental populations and how they affect admixture estimates. The "Middle Eastern" component estimate will increase if central and eastern Europeans are used as representative of the European admixture, while the "European" estimate will increase if Italians are used. But, the same applies to the other end of the continuum: if ancestral Jews were indeed like current Middle Easterners such as the Druze or Palestinians, but the latter may have moved (in genetic space) away from ancient Levantines due to subsequent admixture (Arabs, and in the case of Palestinians even Africans): this would reduce the inferred Middle Eastern component.
Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population
Steven M. Bray et al.
The Ashkenazi Jewish (AJ) population has long been viewed as a genetic isolate, yet it is still unclear how population bottlenecks, admixture, or positive selection contribute to its genetic structure. Here we analyzed a large AJ cohort and found higher linkage disequilibrium (LD) and identity-by-descent relative to Europeans, as expected for an isolate. However, paradoxically we also found higher genetic diversity, a sign of an older or more admixed population but not of a long-term isolate. Recent reports have reaffirmed that the AJ population has a common Middle Eastern origin with other Jewish Diaspora populations, but also suggest that the AJ population, compared with other Jews, has had the most European admixture. Our analysis indeed revealed higher European admixture than predicted from previous Y-chromosome analyses. Moreover, we also show that admixture directly correlates with high LD, suggesting that admixture has increased both genetic diversity and LD in the AJ population. Additionally, we applied extended haplotype tests to determine whether positive selection can account for the level of AJ-prevalent diseases. We identified genomic regions under selection that account for lactose and alcohol tolerance, and although we found evidence for positive selection at some AJ-prevalent disease loci, the higher incidence of the majority of these diseases is likely the result of genetic drift following a bottleneck. Thus, the AJ population shows evidence of past founding events; however, admixture and selection have also strongly influenced its current genetic makeup.
Link
July 14, 2010
mtDNA of Yemeni and Ethiopian Jews
Mitochondrial DNA analysis also revealed a high diversity of sub-Saharan African and Eurasian haplotypes in both the Yemenite and Ethiopian Jewish populations (see Fig. 2). Specifically, common haplotypes (haplotypes present at [5%) in Yemenite Jews include the African haplogroup L3x1 and Eurasian haplogroups R0a (renamed from (preHV)1 (Torroni et al., 2006), HV1, J2a1a [renamed from J1b (Palanichamy et al., 2004)] K, R2, U, and U1, and in Ethiopian Jews include African haplogroups L2a1b2 and L5a1 and Eurasian haplogroups R0a and M1a1 (see Fig. 2). Overall, sub-I think that the authors' conclusion that Yemenite Jews are partially descended from Israeli exiles is premature. Sure, they can exclude large-scale introgression of Yemeni mtDNA, but the universe of possibilities is not limited to either Israeli or Yemenite.
Saharan African L haplotypes [hereafter referred to as L(xM,N), i.e., all African haplotypes except M and N, following the nomenclature of Behar et al. (2008)], comprise a large proportion of the genetic variation in both Jewish populations, representing 20% in the Yemenite Jews and 50% in Ethiopian Jews. This high frequency contrasts with other Jewish populations, such as Near Eastern and Ashkenazi Jews, who almost entirely lack L(xM,N) haplogroups (Thomas et al., 2002; Richards et al., 2003).
American Journal of Physical Anthropology doi: 10.1002/ajpa.21360
Mitochondrial DNA reveals distinct evolutionary histories for Jewish populations in Yemen and Ethiopia
Amy L. Non et al.
Abstract
Southern Arabia and the Horn of Africa are important geographic centers for the study of human population history because a great deal of migration has characterized these regions since the first emergence of humans out of Africa. Analysis of Jewish groups provides a unique opportunity to investigate more recent population histories in this area. Mitochondrial DNA is used to investigate the maternal evolutionary history and can be combined with historical and linguistic data to test various population histories. In this study, we assay mitochondrial control region DNA sequence and diagnostic coding variants in Yemenite (n = 45) and Ethiopian (n = 41) Jewish populations, as well as in neighboring non-Jewish Yemeni (n = 50) and Ethiopian (previously published Semitic speakers) populations. We investigate their population histories through a comparison of haplogroup distributions and phylogenetic networks. A high frequency of sub-Saharan African L haplogroups was found in both Jewish populations, indicating a significant African maternal contribution unlike other Jewish Diaspora populations. However, no identical haplotypes were shared between the Yemenite and Ethiopian Jewish populations, suggesting very little gene flow between the populations and potentially distinct maternal population histories. These new data are also used to investigate alternate population histories in the context of historical and linguistic data. Specifically, Yemenite Jewish mitochondrial diversity reflects potential descent from ancient Israeli exiles and shared African and Middle Eastern ancestry with little evidence for large-scale conversion of local Yemeni. In contrast, the Ethiopian Jewish population appears to be a subset of the larger Ethiopian population suggesting descent primarily through conversion of local women.
Link
June 09, 2010
Genome-wide structure of Jews (Behar et al. 2010)
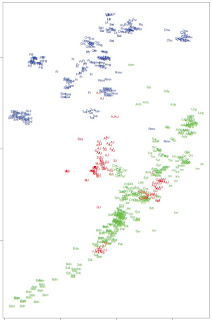 On the left, PCA from Supplementary Figure 3, shows clearly at least three different Jewish clusters. Note the main Ashkenazi/Sephardi cluster halfway between Tuscans and Near Eastern populations, a Yemeni Jewish cluster coinciding with Bedouins and Saudi Arabians, and a West Asian cluster encompassing Georgian, Iranian, Iraqi, etc. Jews.
On the left, PCA from Supplementary Figure 3, shows clearly at least three different Jewish clusters. Note the main Ashkenazi/Sephardi cluster halfway between Tuscans and Near Eastern populations, a Yemeni Jewish cluster coinciding with Bedouins and Saudi Arabians, and a West Asian cluster encompassing Georgian, Iranian, Iraqi, etc. Jews.Below is ADMIXTURE analysis in the global context.
There is a ton of information in the above figure, for Jews and non-Jews alike. Some observations:
- Ethiopians and Ethiopian Jews look identical, between Sub-Saharan Africans and West Asians .
- Sub-Saharan admixture in Egyptians and Yemenites is quite evident; lack of such admixture in Europe and non-Arabs from West Asia.
- A little Caucasoid admixture in Mongols
- Split of Mongoloids into two clusters, which appear to be "northern" and "southern"
- Central Asian Turkic speakers (Uygur, Uzbek) derived from both Mongoloid sub-clusters; their Caucasoid components are mainly West Asian (light green) rather than north European (dark blue)
- Non-European components in Russians are resolved into Caucasoid light green and "north Mongoloid" (see above)
- A little of the "north Mongoloid" component in Turks and some populations from the Caucasus, not much elsewhere in West Eurasia
- South Asian (green) component in Cambodians
- Russians and Lithuanians lack south European (light blue) component but have some west Asian (light green)
- Cypriots are split between West Asia and Southern European components, with minority Semitic (Phoenicians or Syrian Christians?) and northern European ones.
- French Basque and Sardinians lack West Asian component (light green)
This study further uncovers genetic structure that partitions most Jewish samples into Ashkenazi–north African– Sephardi, Caucasus–Middle Eastern, and Yemenite subclusters (Fig. 2). There are several mutually compatible explanations for the observed pattern: a splintering of Jewish populations in the early Diaspora period, an underappreciated level of contact between members of each of these subclusters, and low levels of admixture with Diaspora host populations.
The genome-wide structure of the Jewish people
Doron M. Behar et al.
Contemporary Jews comprise an aggregate of ethno-religious communities whose worldwide members identify with each other through various shared religious, historical and cultural traditions1, 2. Historical evidence suggests common origins in the Middle East, followed by migrations leading to the establishment of communities of Jews in Europe, Africa and Asia, in what is termed the Jewish Diaspora3, 4, 5. This complex demographic history imposes special challenges in attempting to address the genetic structure of the Jewish people6. Although many genetic studies have shed light on Jewish origins and on diseases prevalent among Jewish communities, including studies focusing on uniparentally and biparentally inherited markers7, 8, 9,10, 11, 12, 13, 14, 15, 16, genome-wide patterns of variation across the vast geographic span of Jewish Diaspora communities and their respective neighbours have yet to be addressed. Here we use high-density bead arrays to genotype individuals from 14 Jewish Diaspora communities and compare these patterns of genome-wide diversity with those from 69 Old World non-Jewish populations, of which 25 have not previously been reported. These samples were carefully chosen to provide comprehensive comparisons between Jewish and non-Jewish populations in the Diaspora, as well as with non-Jewish populations from the Middle East and north Africa. Principal component and structure-like analyses identify previously unrecognized genetic substructure within the Middle East. Most Jewish samples form a remarkably tight subcluster that overlies Druze and Cypriot samples but not samples from other Levantine populations or paired Diaspora host populations. In contrast, Ethiopian Jews (Beta Israel) and Indian Jews (Bene Israel and Cochini) cluster with neighbouring autochthonous populations in Ethiopia and western India, respectively, despite a clear paternal link between the Bene Israel and the Levant. These results cast light on the variegated genetic architecture of the Middle East, and trace the origins of most Jewish Diaspora communities to the Levant.
Link
June 03, 2010
Two major groups of living Jews (Atzmon et al. 2010)
Next, each of 2407 European subjects was assigned into one of 10 groups based on geographic region: South:Italy, Swiss-Italian; Southeast: Albania, Bosnia-Herzegovina, Bulgaria, Croatia, Greece, Kosovo, Macedonia, Romania, Serbia,Slovenia, Yugoslavia; Southwest: Portugal, Spain; East: CzechRepublic, Hungary; East-Southeast: Cyprus, Turkey; Central:Austria, Germany, Netherlands, Swiss-German; West: Belgium,France, Swiss-French, Switzerland; North: Denmark, Norway,Sweden; Northeast: Finland, Latvia, Poland, Russia, Ukraine;Northwest: Ireland, Scotland, UK.
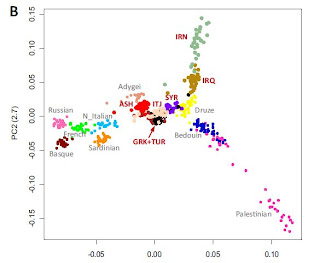

Admixture with local populations, including Khazars and Slavs, may have occurred subsequently during the 1000 year (2nd millennium) history of the European Jews. Based on analysis of Y chromosomal polymorphisms, Hammer estimated that the rate might have been as high as 0.5% per generation or 12.5% cumulatively (a figure derived from Motulsky), although this calculation might have underestimated the influx of European Y chromosomes during the initial formation of European Jewry. Notably, up to 50% of Ashkenazi Jewish Y chromosomal haplogroups (E3b, G, J1, and Q) are of Middle Eastern origin,15 whereas the other prevalent haplogroups (J2, R1a1, R1b) may be representative of the early European admixture. The 7.5% prevalence of the R1a1 haplogroup among Ashkenazi Jews has been interpreted as a possible marker for Slavic or Khazar admixture because this haplogroup is very common among Ukrainians (where it was thought to have originated), Russians, and Sorbs, as well as among Central Asian populations, although the admixture may have occurred with Ukrainians, Poles, or Russians, rather than Khazars. In support of the ancestry observations reported in the current study, the major distinguishing feature between Ashkenazi and Middle Eastern Jewish Y chromosomes was the absence of European haplogroups in Middle Eastern Jewish populations.
AJHG doi:10.1016/j.ajhg.2010.04.015
Abraham's Children in the Genome Era: Major Jewish Diaspora Populations Comprise Distinct Genetic Clusters with Shared Middle Eastern Ancestry
Gil Atzmon et al.
Abstract
For more than a century, Jews and non-Jews alike have tried to define the relatedness of contemporary Jewish people. Previous genetic studies of blood group and serum markers suggested that Jewish groups had Middle Eastern origin with greater genetic similarity between paired Jewish populations. However, these and successor studies of monoallelic Y chromosomal and mitochondrial genetic markers did not resolve the issues of within and between-group Jewish genetic identity. Here, genome-wide analysis of seven Jewish groups (Iranian, Iraqi, Syrian, Italian, Turkish, Greek, and Ashkenazi) and comparison with non-Jewish groups demonstrated distinctive Jewish population clusters, each with shared Middle Eastern ancestry, proximity to contemporary Middle Eastern populations, and variable degrees of European and North African admixture. Two major groups were identified by principal component, phylogenetic, and identity by descent (IBD) analysis: Middle Eastern Jews and European/Syrian Jews. The IBD segment sharing and the proximity of European Jews to each other and to southern European populations suggested similar origins for European Jewry and refuted large-scale genetic contributions of Central and Eastern European and Slavic populations to the formation of Ashkenazi Jewry. Rapid decay of IBD in Ashkenazi Jewish genomes was consistent with a severe bottleneck followed by large expansion, such as occurred with the so-called demographic miracle of population expansion from 50,000 people at the beginning of the 15th century to 5,000,000 people at the beginning of the 19th century. Thus, this study demonstrates that European/Syrian and Middle Eastern Jews represent a series of geographical isolates or clusters woven together by shared IBD genetic threads.





















