 The paper is behind a paywall, but there is plentiful raw genetic data in the online supplement. I'll probably have much more to say on this when I read it, but here's the groundbreaking part:
The paper is behind a paywall, but there is plentiful raw genetic data in the online supplement. I'll probably have much more to say on this when I read it, but here's the groundbreaking part:Most of this sample belonged to haplogroup G2a-P15 with some I2a-P37.2 also represented.
G2a was also one of the haplogroups represented in a small sample from Neolithic Central Europe. I think we can now safely say that G2a may have been the main Neolithic link that ties the farmers that went north across the Balkans to Central Europe, and those that followed the western, maritime route to the Western Mediterranean. The unambiguous West Asian origin of this lineage should put to rest any ideas about Neolithic farmers in the Western Mediterranean being descended from indigenous Mesolithic foragers.
I-P37.2 is also quite interesting, as it is tied to the Balkans, but also modern Southwestern Europe (it is especially frequent in Sardinia in its derived M26+ form). ISOGG tells me that:
I2-M438 et al includes I2* which shows some membership from Armenia, Georgia and Turkey; I2a-P37.2, which is the most common form in the Balkans and Sardinia. I2a1-M26 is especially prevalent in Sardinia. I2b-M436 et al reaches its highest frequency along the northwest coast of continental Europe. I2b1-M223 et al occurs in Britain and northwest continental Europe. I2b1a-M284 occurs almost exclusively in Britain, so it apparently originated there and has probably been present for thousands of years.
If these aren't signals of a maritime pioneer colonization that followed the maritime route along the Mediterranean and Atlantic, I don't know what is.

 The Y-chromosome is an extreme outlier compared to modern groups, probably because of its heavy G2a domination, whereas the mtDNA from Treilles appears just like a normal and unexceptional Mediterranean-type population.
The Y-chromosome is an extreme outlier compared to modern groups, probably because of its heavy G2a domination, whereas the mtDNA from Treilles appears just like a normal and unexceptional Mediterranean-type population.
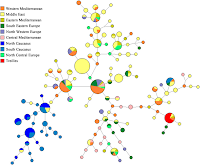
UPDATE V: The G2a median joining network shows that the Treilles haplotypes are disjoint from those that dominate the North Caucasus, with clear links to the Middle East, Central/East Mediterranean regions, as well as the South Caucasus.
Ancient DNA reveals male diffusion through the Neolithic Mediterranean route
Marie Lacan et al.
The Neolithic is a key period in the history of the European settlement. Although archaeological and present-day genetic data suggest several hypotheses regarding the human migration patterns at this period, validation of these hypotheses with the use of ancient genetic data has been limited. In this context, we studied DNA extracted from 53 individuals buried in a necropolis used by a French local community 5,000 y ago. The relatively good DNA preservation of the samples allowed us to obtain autosomal, Y-chromosomal, and/or mtDNA data for 29 of the 53 samples studied. From these datasets, we established close parental relationships within the necropolis and determined maternal and paternal lineages as well as the absence of an allele associated with lactase persistence, probably carried by Neolithic cultures of central Europe. Our study provides an integrative view of the genetic past in southern France at the end of the Neolithic period. Furthermore, the Y-haplotype lineages characterized and the study of their current repartition in European populations confirm a greater influence of the Mediterranean than the Central European route in the peopling of southern Europe during the Neolithic transition.
Link
What is absent is also quite interesting as what is present. The absence of E1b1b is consistent with my theory about the Bronze Age Greek expansion of that haplogroup in Europe that has been tied to the historical Greeks of the West Mediterranean.
R-M269 which, because of its apparent young Y-STR age has been tied by some to either the Mediterranean or Central European Neolithic is conspicuous absently from both at the moment. It may yet surface in a Neolithic context, but its absence this late from a region where, today, it is abundant only adds to its mystery. The absence of J2 is equally mysterious, as this is another putative Neolithic lineage which has failed to appear so far in a Neolithic context, while its J1 sister clade did make an appearance in much later aboriginals from the Canary Islands.
UPDATE I: Interestingly, some French researchers had noted a littoral distribution of haplogroups I, J, G in the Finistère, on the Atlantic side.
UPDATE II: I was reviewing my Ancient Y-chromosome studies compendium and one thing starts to become clear: how many of the earliest samples we have were dominated by 1-2 haplogroups, whereas there is a plethora of haplogroups in most modern populations: Treilles, Krasnoyarsk, Xiaohe, Pengyang all belonged to a single haplogroup, while Yangtze China to several lineages, all of which were in the O haplogroup.
Look at the MDS plot of the Y-chromosome and mtDNA from Treilles:

 The Y-chromosome is an extreme outlier compared to modern groups, probably because of its heavy G2a domination, whereas the mtDNA from Treilles appears just like a normal and unexceptional Mediterranean-type population.
The Y-chromosome is an extreme outlier compared to modern groups, probably because of its heavy G2a domination, whereas the mtDNA from Treilles appears just like a normal and unexceptional Mediterranean-type population.Perhaps the modern Caucasus where particular ethnic groups are dominated by particular Y-haplogroups is a good analogy for prehistoric man, with many different groups with their signature haplogroups kept disjoint patrilineal gene pools before beginning to merge in late prehistorical and historical times.
UPDATE III: A poster at dna-forums as well as Ken Nordtvedt both agree that the I2a haplotypes belong to I-M26, a haplogroup that is modal in the SW Mediterranean, reaching very high frequencies in Sardinia. This may be consistent with the great biological continuity since the Neolithic in Sardinia, continuity which is also evident on the mtDNA. It also shows why the inference of pre-Neolithic I2a in Sardinia was flawed because of the use of the evolutionary mutation rate, while the origin and expansion of I-M26 in "genealogical rate" years becomes 5-7ky, consistent with the Neolithic origin of that haplogroup and the ancient DNA presence in Neolithic France.
UPDATE IV: Table S4 lists (in %) shared mtDNA lineages between Treilles and modern populations. The top ones are: Welsh (17.391), Cornish (16.667), Central Greeks (14.286), Bulgarians (12.5). Several Italian groups as well as South Tyrol Ladins and Germans are also greater than 10%.

UPDATE V: The G2a median joining network shows that the Treilles haplotypes are disjoint from those that dominate the North Caucasus, with clear links to the Middle East, Central/East Mediterranean regions, as well as the South Caucasus.
UPDATE VI: Some more good news: "The ancient DNA Lacan is now extracting from skeletons across France and Spain, Haak says, should provide more “piece[s] of the enormous puzzle we are trying to put together.”
PNAS doi: 10.1073/pnas.1100723108
Ancient DNA reveals male diffusion through the Neolithic Mediterranean route
Marie Lacan et al.
The Neolithic is a key period in the history of the European settlement. Although archaeological and present-day genetic data suggest several hypotheses regarding the human migration patterns at this period, validation of these hypotheses with the use of ancient genetic data has been limited. In this context, we studied DNA extracted from 53 individuals buried in a necropolis used by a French local community 5,000 y ago. The relatively good DNA preservation of the samples allowed us to obtain autosomal, Y-chromosomal, and/or mtDNA data for 29 of the 53 samples studied. From these datasets, we established close parental relationships within the necropolis and determined maternal and paternal lineages as well as the absence of an allele associated with lactase persistence, probably carried by Neolithic cultures of central Europe. Our study provides an integrative view of the genetic past in southern France at the end of the Neolithic period. Furthermore, the Y-haplotype lineages characterized and the study of their current repartition in European populations confirm a greater influence of the Mediterranean than the Central European route in the peopling of southern Europe during the Neolithic transition.
Link

84 comments:
"The unambiguous West Asian origin of this lineage should put to rest any ideas about Neolithic farmers in the Western Mediterranean being descended from indigenous Mesolithic foragers."
To the extent that almost all modern humans in Europe made their way there via ancestors in West Asia, I don't agree that this tells us very much. The mtDNA hg disconnect in the ancient DNA is far more powerful on this score.
The points on the absence of R-M269, J2 and E1b1b are well taken. The absence of R-M269 is particularly striking because it is so common now and is the missing hg most frequently inferred based on modern geographic distribution to be Epipaleolithic.
The point on absence of hg diversity in ancient samples is also well taken. Apparently Hebrew Bible type scenarios of patrilineal tribes expanding by natural growth rather than assimilation were meaningfully accurate for a long time.
"R-M269 which, because of its apparent young Y-STR age has been tied by some to either the Mediterranean or Central European Neolithic is conspicuous absently from both at the moment. It may yet surface in a Neolithic context, but its absence this late from a region where, today, it is abundant only adds to its mystery."
I don't see a mystery. The amateur estimates using actual mutation rates put the spread of R1b into Western Europe clearly post-Neolithic. R1b in Western Europe is in all likelihood associated with the dispersal of Indo-European.
The mystery isn't so much about its age (the combination of the Y-STR evidence and the lack of it in many early samples are pretty convincing) but about its origins.
Some have argued that it came to Europe from the steppes (north of the Black Sea), but so far it's absent in Corded Ware culture and in the eastern steppe. Others have argued it came from West Asia (south of the Black Sea), based largely on phylogenetic considerations and modern Y-STR variance, but these don't seem particularly safe after so many ancient DNA surprises.
"I don't see a mystery. The amateur estimates using actual mutation rates put the spread of R1b into Western Europe clearly post-Neolithic. R1b in Western Europe is in all likelihood associated with the dispersal of Indo-European."
That's very unlikely, in my opinion. First, look at the Basques: they don't speak an IE-language, yet they have one of the highest % of R1b among Europeans. On the other hand it's quite difficult to believe that 80% of western European men trace their lineages to migrations that occurred less than 5.000 years ago.
Why do you keep talking about the EXPANSION of E-V13 to other parts of Europe and not its age? Could it not have arisen in Greece or Anatolia as far back as the Mesolithic or do you dismiss that speculation?
What about the age of V13?
Is it possible that the people who brought R1b to Europe also brought lactase persistence with them? This might explain why R1b is so common in modern western Europe even though it appears to be very young.
If G2 were a major marker of neolithic expansion from the east then what is clear is that it did not take. G2 is a minor haplogroup in Europe today. Even in France (2.5%). No evidence for Neolithic extermination here.
It seems to me this is a just a G2 dominated kinship group. An artifact of a patrilocal society. More likely to have originated in Switzerland (10%) than Turkey (11%).
The next village over was probabaly dominated by a completely different haplogroup.
The mitochondrial DNA is interesting too and worth posting.
H1/H3 already dominates so that expansion has clearly already occurred in southern France, as expected if this is a post glacial expansion. Patterns for these haplogroups have previously indicated an expansion from Spain.
U5 is very well represented so clearly paleolithic women were a significant part of this early "neolithic" society.
No typical neolithic mitochondrial haplogroups observed so if Y haplogroup G2 came from the east in the neolithic they did not bring significant numbers of women with them.
The authors match the ancient G2 most closely with a modern population in central Spain. This would normally suggest central Spain as the immediate source (as done in other papers of this type). But in this paper this fact is ignored. I am sure it would not have been ignored if the most similar region had been central Turkey. G2 is very high in Cantabria in modern populations.
The very fact that G2a is present in France 5,000 years ago elminates the Alans as the source of this haplogroup in Europe. Clearly they were present well before the Alans, Romans etc.
In Summary, a patrilocal G2a kinship group (likely mostly descended from 1 man) connected to central Spain, including matrilinear lines from Spain (or at least the Pyrenean refuge) and matrilinear lines known to be typically paleolithic. This early neolithic community is at least 50% (female) local assimulation. And the G2a connections to central Spain are intriguing.
"That's very unlikely, in my opinion. First, look at the Basques: they don't speak an IE-language, yet they have one of the highest % of R1b among Europeans. On the other hand it's quite difficult to believe that 80% of western European men trace their lineages to migrations that occurred less than 5.000 years ago."
Could we interpret the presence of R1b in Basques as resulting from gene flow from Indo-Europeans? I understand that its frequency in Basques is high but then Basques are surrounded by a sea of IE speakers.
This is the late Neolithic(Æneolithic) or more popularly called Copper Age we're talking about really--an era of considerable commerce and mobility.
The Neolithic timeframe runs from 5-15,000 years ago. The Mesolithic time is rather an arbitrary and a matter of semantics argumentation besides so I dont think this means a whole lot or covers the entire "Neolithic".
"Why do you keep talking about the EXPANSION of E-V13 to other parts of Europe and not its age? Could it not have arisen in Greece or Anatolia as far back as the Mesolithic or do you dismiss that speculation?"
I think it's unlikely. Maju and I have just finished a long to and fro concerning E1b1b1 and I think we now agree that the haplogroup entered the Mediterranean, probably via the Nile, during the early Holocene, perhaps around 10,000 years ago.
@Average Joe:
Lactase persistence concentrates in the north West of Europe. British Isles, Benelux, Scandinavia and Germany.
Spain is full of R1b people too, but has relatively few (for European standarts) people with Lactose tolerance. Even Poland has more (wich is R1a dominated with only a handfull R1b), on some maps even Finland, northern Russia and the Baltic states are more Lactose tolerant than Spanish.
So, R1b alone does not do the trick.
Its however something "North European". Specially "North West European".
Spain is full of R1b people too, but has relatively few (for European standarts) people with Lactose tolerance
Could you post some links supporting this? I have not been able to find any information that backs up your claim that lactose tolerance is low in Spain compared to other European countries.
"Could you post some links supporting this? I have not been able to find any information that backs up your claim that lactose tolerance is low in Spain compared to other European countries."
Several typical maps:
http://www.dsm.com/le/static/maxilact/images/lactose_intolerance.jpg
http://www.naturheilpraxis-bornemann.de/uploads/pics/lactose_intolleranz_gesamtbevoelkerung_gross.gif
http://www.wirklich.ch/milchzucker/lactoseintoleranz-map.jpg
http://www.packaleks.de/images/li_globe_small.svg.png
http://demo.decodeme.com/images/trait/li/header.jpg
http://1.bp.blogspot.com/_Ish7688voT0/S3pd_AUYorI/AAAAAAAACS4/c_rFUFT9dlI/s1600-h/lpp.png
Its however something "North European". Specially "North West European".
In the sense of "most frequent in Northern Europe", yes.
In the sense of "originated in North Europe", no, as it the allele that has reached a high frequency in North Europeans is far too widespread.
"Could we interpret the presence of R1b in Basques as resulting from gene flow from Indo-Europeans? I understand that its frequency in Basques is high but then Basques are surrounded by a sea of IE speakers."
It's highly unlikely they were replaced by I.E. speakers, but still, they don't speak any IE tongue. The authors didn't sample any North Africans. There are many genetic similarities between North Africans and Europeans which surely date much older than 5.000 years ago. I'd be interesting to see if they're as related to Treilles as any other European population or not.
"More likely to have originated in Switzerland (10%) than Turkey (11%)"
--Don't know about 10%; but could this be Raetic; a possible Etruscan relation? -- And a later migration, anyway.
"In the sense of "originated in North Europe", no, as it the allele that has reached a high frequency in North Europeans is far too widespread."
Possibly.
But I almost bet, this allele aswell as the alleles that cause light eyes and fair hair are part of that allele pattern, that admixture interprets as the "Northern European component".
In Europe, these 4 things (lactose tolerance, blue eyes, fair hair, northern European aDNA component) roughly cover the same space, only with different distribution centers (hair/eyes/Northern aDNA component all huddle around the baltic sea, spreading towards the North Sea.
And the Lactose tolerance huddles around the Northsea and spreads towards the Baltic Sea.
@Terry: no we do not agree. That I stopped replying to you indicates boredom and annoyance, not agreement.
@Average Joe: actually Spaniards are quite good at digesting raw milk for global and even European standards (cf. Wikipedia, where a host of reference and links can be followed - though the map is absolutely misleading).
Non-Basque Spaniards have on average only 15% lactose intolerance following Wikipedia. Basques have the lowest lactose intolerance worldwide (0.3%), followed by the Dutch (1%), etc.
However "South French" (which "South French" exactly, I could not find out but I presume that SE French, after considering Basque figures) have a much higher level: 65% intolerant, quite extreme for European levels.
As Annie pointed out (the only one making any sense in this joint), there's only 2 y-dna samples, an I2a and a G2a, the other 20 y-dna samples are inbreeding repeats of those first 2. The Admin of the G Project has said that the haplotype presented for G could be any subgroup of G, they're all impossible to tell apart with the yhrd set of STR markers selected. The I2a fits the modal of I-M26 like a glove, absolutely perfectly, I can even determine which specific subcluster of I-M26 it belongs to. I2a is diffused all over Europe and the Middle East, but I-M26 is virtually exclusive of southwest Europe, being almost impossible to find elsewhere.
Once again, mtdna H is much lower than observed today, in this case just 24%. And once again, U5 is much higher than today, in this case 21%. There was yet another case of U with CRS haplotype. Today, only 1 in 300 U samples has a CRS haplotype, and these rare cases are all in U4 or U8. There was no N1a sample! This made up 25% of Haak's 2005 Neolithic samples. It also made up 1 of 3 of a recent Neolithic mtdna study of France. Remember that other recent mtdna study of Denmark? Most were historic, but they tested 3 Neolithic samples. All 3 were U5.
Dienekes,
"Some have argued that it came to Europe from the steppes (north of the Black Sea), but so far it's absent in Corded Ware culture and in the eastern steppe."
I don't think this is unexpected.
(1) I believe only a handful of Corded Ware Y results have been published.
(2) The R1b lineage that now dominates Western Europe was starting from 0 and riding west in an expanding population.
(3) The earliest R1b so far documented comes from Bronze Age Germany, where it was found with a larger number of R1a.
Neandertalerin,
"First, look at the Basques: they don't speak an IE-language, yet they have one of the highest % of R1b among Europeans. On the other hand it's quite difficult to believe that 80% of western European men trace their lineages to migrations that occurred less than 5.000 years ago."
Regardless of how easy it is for you to believe, that's what the evidence points to. Basque R1b also has limited diversity even within Western European R1b: I believe Basque R1b is limited to R-P312, while R-P312, R-U106, and occasional more divergent R1b lineages are found among Western European IE speakers. Given the young age and present distribution of R1b, and the relative numbers of IE speakers and Basque speakers, it's reasonable to suppose Basques acquired R1b from IE speakers, while the reverse is not reasonable.
Fanty:
Thanks for the links. Much appreciated. Lactase persistence may not have been brought to Europe by R1b carriers but may have originated in a R1b population that lived in northern Europe.
"... it's reasonable to suppose Basques acquired R1b from IE speakers, while the reverse is not reasonable".
Why not? If IEs would have invaded Basques in figures of 90% (of males), Basques would not be Basques but IE and would speak some sort of IE.
However if (some) IEs are "Basques" who have lost their language and acquired some lesser "IE" genetic admixture, then... well, we get today's Western Europe.
...
Besides that, as I joined this conversation, I want to say that we are before a "collective" (clannic) burial and hence it's absolutely normal that all share one lineage because they all should have shared the same ancestor (I imagine that the I2a types are a from an adopted or illegitimate branch). What this study clarifies is that this shared ancestor was male and hence the culture was already patrilineal and patrilocal.
Maju,
It's not 2003 anymore. Basques are not Paleolithic remnants. R1b is not of great antiquity in W. Europe.
Your argument (as specifically concerns "Basques") wasn't strong then, but it certainly doesn't work now. It would require positing a completely unnecessary, unsubstantiated, fleeting conquest of W. Europe by Basques.
A small amount of gene flow compounded over a couple thousand years can have dramatic effects.
I think it's possible this Treilles settlement (a relatively late Neolithic colony) represents a secondary wave after a first river valley occupying, R1b-rich one and before a later J2, E1b1b-rich secondary wave associated in the Near East with the Semitic language family expansion.
High levels of U5 mtDNA can be interpreted as indicating a new agricultural settlement in previously fallow land not productive enough for the first wave but good enough for a better equiped secondary wave.
So R1bs could be waiting to be discovered in nearby older settlements and possibly in late hybridized forager populations.
I speculated before major agricultural innovations with large surpluses could produce both expansions into neighbouring lands occupyied by less advanced agriculturalists, and local demographic expansions with larger elite-formation in the old centre.
I'd be interested to see y-haplogroup studies on pre-Akkadian Levantine and Mesopotamian remains.
I feel these postulated G-rich 3000bc European Neolithic populations may well correspond to a "West Asian" (or "EMPC") component in ADMIXTURE (G2a is also more prevalent in Northern Indians and Upper-Caste Southern Indians).
J2 is not associated with the spread of Semitic languages, as it occurs in India/Central Asia where Semitic languages are not attested with a lack of the "Southwest Asian" autosomal component that is important in Semitic groups.
J2 also happens to differentiate Germans from Poles
http://dienekes.blogspot.com/2005/06/strong-differentiation-between-germans.html
J2 and R1b seem correlated in Germans:
http://dienekes.blogspot.com/2008/04/haplogroup-correlations-in-germans.html
They also seem to be correlated in Southeastern Europe:
http://dienekes.blogspot.com/2005/08/haplogroup-frequency-correlations-in.html
The most likely source for R1b in Europe is West Asia where J2 is also frequent.
I increasingly believe that J2+R1b spread into Europe, and perhaps a founder effect in the Balkans is responsible for the apparent youth of R1b in most of Europe; in the Balkans R1b appears only a little younger than in West Asia.
The spread of R1a1a-M17 is also quite puzzling due to its twin foci in eastern Europe and south Asia, with the added problem of the apparent presence of all R*, R1*, R1a*, R1a1* in the Near East.
Speaking of the Basques, the most interesting result from the Dodecad Project is that they seem to lack the "West Asian" component than their French/Spanish neighbors possess. J2 was also not found in an ancient Basque cemetary.
I would say that a good hypothesis is that J2+R1b groups moved into Europe after the initial Neolithic and up to the Bronze Age; these need not have been homogeneous in culture or language, and a few of them escaped Indo-Europeanization, such as the Basques and various other non-IE groups attested for Western and Southwestern Europe.
Dienekes continues to not acknowledge that the y-dna consists of only 2 samples. There's no doubt about the matter. This is the result of patrilocality and it was observed in a previous much discussed ancient y-dna study of Germany, where all the lineages belonged to a rare branch of y-dna I.
I have to reiterate that not only does the I2a sample belong to M26, but it matches almost perfectly one of its subclusters, the most common one. There are 2 main subclusters of M26, the lesser has 391=9, the more important one has 391=10. The latter is the one this ancient sample belongs to. The TMRCA of either cluster is 2000 to 2500 years ago.
apparent youth of R1b in most of Europe
R1b in Europe has an age of 3500 years. That's a typical TMRCA age estimate for most haplogroup anywhere in the world. A young age would be 2000 years ago. I don't think any cluster has ever had a TMRCA calculation greater than 5000 years ago.
Dienekes continues to not acknowledge that the y-dna consists of only 2 samples. There's no doubt about the matter. This is the result of patrilocality and it was observed in a previous much discussed ancient y-dna study of Germany, where all the lineages belonged to a rare branch of y-dna I.
Patrilocality in no way detracts from the fact that a sample from a rare haplogroup (today) was found in two different Neolithic sites in Europe.
That's no accident, and while there is plenty of statistical room for the presence of other haplogroups in the early Neolithic, the evidence is clear that G2a was a major Y-haplogroup associated with it.
I don't think any cluster has ever had a TMRCA calculation greater than 5000 years ago.
That's not true. If you look at the data from Cinnioglu et al. (2004), for example, and correct their ages by dividing by 3, there are many haplogroups with an age greater than 5000 years ago.
Speaking of the Basques, the most interesting result from the Dodecad Project is that they seem to lack the "West Asian" component than their French/Spanish neighbors possess. J2 was also not found in an ancient Basque cemetary.
Yes, based on the available genetic data I see no reason to think that Basques have any Indo-European admixture. In fact, they are one of the genetically purest populations in the world.
"A small amount of gene flow compounded over a couple thousand years can have dramatic effects".
That's an unsustainable claim. You'd need a source and there is no one source with so much R1b1b2a1a2 (S116 old nomenclature, I haven't yet been able to remember the new one) anywhere around, except maybe Ireland.
There are sublineages that are specifically Basque and others that are specifically sub-Pyrenean (Gascon, Catalan, Basque and such). Of all known basal sublineages of R1b1b2a1a2 most are found around the old Franco-Cantabrian Region, only the "Irish" M259 subclade is a bit off the picture (but it is anyhow found in South France as well, where the origin may lay).
See here for cool maps and to grasp what is R1b in West Europe. There is another Western lineage R1b1b2a1a1 (U106) specific (not strictly so) of NW Europe that I imagine originated in the Rhine-Danube region and/or the Hamburgian-Ahrensburgian-Maglemosean cultural area.
"R1b is not of great antiquity in W. Europe".
That you will have to demonstrate scientifically (TRMCA is not scientific nor demostrated).
I don't know much about haplogroup R1b in Europe, but this map makes me think it had at least a secondary expansion from the Balkans, not from the Ukrainian steppes.
http://dc302.4shared.com/img/2vvQ-f-w/s7/R1b1b2_sub-structure_V2.png
Maju:
Non-Basque Spaniards have on average only 15% lactose intolerance following Wikipedia. Basques have the lowest lactose intolerance worldwide (0.3%), followed by the Dutch (1%), etc.
Thanks for the info. I guess this shows a possible link between R1b and lactase persistence. The question now is when and where lactase persistence first appeared in a R1b population.
"J2 is not associated with the spread of Semitic languages, as it occurs in India/Central Asia where Semitic languages are not attested"
But isn't Dravidian/Elamite considered to be related to Afro-Asiatic?
"in the Balkans R1b appears only a little younger than in West Asia".
There seems to be a long period of drift in the R1b line. Little diversity between R1b1a (somewhere between Anatolia and Afghanistan) until R1b1a2a1a1, where the line splits into a north European R1b1a2a1a1a and a Mediterranean R1b1a2a1a1b. Both lines are found in the UK but presumably reached there by separate routes. did the two start out from the Balkans?
But isn't Dravidian/Elamite considered to be related to Afro-Asiatic?
There is no verified relationship between them.
As for J2, it is too widespread worldwide to be associated with the spread of Semitic languages. Semitic languages aren't attested as spoken languages in almost all of the:
the Anatolian Peninsula, the Armenian Highlands, the Caucasus, the Balkans, the Italian Peninsula and most parts of Iran
all of which are non-Semitic regions of the world where J2 reaches high frequencies.
Dienekes said:
Speaking of the Basques, the most interesting result from the Dodecad Project is that they seem to lack the "West Asian" component than their French/Spanish neighbors possess. J2 was also not found in an ancient Basque cemetary.
As I commented recently my father (DOD098) has no known non-Irish ancestry, and also has NO "West Asian" component, while having 23% of the Basque component - substantially higher than other Irish tested. His ancestry is from the Irish Midlands, but has a grandmother from Kerry in the extreme South West of the country - who may account for the elevated Basque component.
Patrilocality in no way detracts from the fact that a sample from a rare haplogroup (today) was found in two different Neolithic sites in Europe.
G2a is 7% of France, 13% of Switzerland, and 10% of North Italy. It's not rare. The results are interesting, but not conclusive. The 2 G samples in the older study could well have been a single sample, and in fact that's most likely given that the 2 other ancient y-dna studies have effectively shown such to be the case. It's still an unsually high presence of G, but nothing is definitve for now, as there are only 2 samples from this study, and maybe just 1 sample from the other (including the F*, which could have been a technical glitch).
I don't think any cluster has ever had a TMRCA calculation greater than 5000 years ago.
That's not true. If you look at the data from Cinnioglu et al. (2004), for example, and correct their ages by dividing by 3, there are many haplogroups with an age greater than 5000 years ago.
You went out of your way to dig up that crap to prove me wrong. The 2 columns of estimates, showing continous growth and expanding population, produce results such as 18kya and 45kya, for the same haplogroup! Obviously this isn't a variance estimate but some simulated crap. Just take the samples and calculate their variance on your own. It shouldn't take 5 minutes if you already have the xls file from Ken Nordtvedt. I did a quick calculation of some 15 haplogroups and they varied between 2000 and 5000 years ago, with R1b1b2 falling in the middle of the range. Typical results.
The I2a sample of the study is almost a perfect match for the most common cluster of I2a-M26, in every aspect of its haplotype. It's just perfect. Except that this cluster has a TMRCA age of only 2000 years ago, and the sample is from 5000 years ago. LOL
Anyway, your whole argument is almost laughably ridiculous. It seems that people like you think that because farmers may have invaded Europe from Anatolia 8000 years ago, then somehow it opens up the door to subsequent repeats. This is like saying that since Spain destroyed America 500 years ago, it would somehow be possible for them to be able to do the same again today. Presuming it were true that Anatolian farmers invaded and replaced the people of Europe 8000 years ago, then that's it. Now they're farmers, too, with the same technology and density. There isn't going to be another physical replacement. In fact, if anything, 3500 years ago it would be Europeans who would be more able, with much more numbers, to invade and replace the people of Anatolia.
All TMRCA estimates always produce ages of just 2000 to 5000 years. So apparently the world can trace its y-dna to just 2 or 3 dozen men that lived barely 3000 years ago. It's a little more likely that the entire theory of TMRCA is a piece of ssss. And this study has found a sample that wasn't suppose to exist 5000 years ago, hell, not even 2500 years ago.
Anyway, your whole argument is almost laughably ridiculous. It seems that people like you think that because farmers may have invaded Europe from Anatolia 8000 years ago, then somehow it opens up the door to subsequent repeats. This is like saying that since Spain destroyed America 500 years ago, it would somehow be possible for them to be able to do the same again today.
That's a good example, since most of the people who live in the Americas today did not arrive there in 1492 but centuries later, and people kept on arriving.
So, I see absolutely no reason to think that the first Neolithic colonists in any region of the world were the only colonists and gene flow subsequently ceased. Rather, there was a continuous process of migration over millennia.
If sea-borne colonists from West Asia established the earliest Neolithic in the West Mediterranean and thousands of years later Greek colonists from West Asia, and Phoenician colonists from the Levant, established their own colonies in the West Mediterranean, I see absolutely no reason to think that in the intervening years gene flow from the east to the west had ceased.
That's a good example, since most of the people who live in the Americas today did not arrive there in 1492 but centuries later, and people kept on arriving.
But it's not a good example, because your argument is that 3500 +/- 500 years ago a massive unique event, a wave of people flowed into Europe, making a specific genetic change in the y-dna, R1b1b2, whose diversity can be calculated so as to determine when that unique event happened. Even your counter-argument doesn't make sense. Again you just assume that Europe must have always been some nothing land where anybody could set up camp and do what the hell he wants. The excavations of Old Europe, from Romania, Serbia, and Bulgaria, from 6000 years ago, show that at that time this region had the largest cities anywhere in the world, and their pottery and metal workmanship was on a level unparalleled anywhere else, also. Europe wasn't a happy hunting ground where any foreigner could come over and set up shop and bring with him 1 million relatives. The only chance was 8000 years ago. After that, they became farmers, exploded in population and the window of opportunity was closed. Just like if Spain were to try to reconquer Latin America today, an attempt by Anatolians to completely rearrange Europe for a 2nd time some 3500 years ago would have been a massive failure; Europe's population must have been at least 5 times greater than that of Anatolia.
I was refering to an association with Semitic in the Near East not necessarily outside it. Genetics tend to associate with linguistics for such times, I think, but one early linguistic conversion (not a rare phenomenon even in the Americas - early Guarani use by Iberian colonists in SAm) can result in different language families, similar Y-genetics downstream.
As for R1b-J2 correlation, if J2 arrived in European R1b-rich regions wouldn't this wave "pick up" a lot of also agriculturalist R1b as it travelled.
Indian J2 is definitely interesting. Don't know why. Maybe merchant elite relations? Very old trade links with NE (back to Sumerian times presumably, then Ptolemaic Egypt- maybe earlier but undocumented, with Arabia).
As for Basques and Sardinians, I think that may be the same main dish with very different "sauces".
I'm just speculating but South Mesopotamian, Western Iranian plateau R percursors? (close to both Anatolia and South Caucasus, with early expansion into India, later mostly swamped in homeland?).
Maju,
You're fairly confused about the current state of the R1b tree.
http://www.u152.org/images/stories/Draft_P312_Tree_v005.png
SRY2627 is a subclade L176.2, which has a wide distribution and no indication of an origin in SW France or Iberia. The same can be said for SRY2627 itself, for that matter.
M153 shares a recently discovered basal subclade (Z196) within R-P312 with L176.2.
Of the currently categorized basal subclades of R-P312, none that I know of show evidence of origins in SW France or Iberia. U152 and L21 show their greatest diversity in other regions of France and/or Germany. I believe L238 is associated with Scandinavia, and the little that I know of DF19 suggests it's more common in Northern Europe as well.
"That's an unsustainable claim. You'd need a source and there is no one source with so much R1b1b2a1a2 (S116 old nomenclature, I haven't yet been able to remember the new one) anywhere around, except maybe Ireland."
There's this thing called genetic drift.
"H1/H3 already dominates so that expansion has clearly already occurred in southern France, as expected if this is a post glacial expansion. Patterns for these haplogroups have previously indicated an expansion from Spain.
U5 is very well represented so clearly paleolithic women were a significant part of this early "neolithic" society."
Very interesting. Given the U5 is dominant in very old mtDNA samples from which H1/H3 are virtually absent, and its pretty rare now, while H1/H3 are much more common now, 5000 years ago has to very close to the midpoint of the transition in mtDNA hg frequency. And in a simple two hg pool model, they should following something approaching a logistic curve, in which the balance point between one group of hgs being dominant and the other group of hgs being dominant tends to be very brief while the tails on either end tend to be quite long.
This would be a moment a bit like capturing the population genetics of Mexico in the late 1500s or early 1600s when it was a midway between the pre-Columbian mix and the modern one.
The case that this is capturing admixture and assimilation with a foreign biased male population in a patrilocal/patrlineal society and a local biased female population at a time close in time to when it started to happen also seems strong and is exciting given the lack of clarity about the extent to which this was happening (and it may represent a difference between the Mediterranean/Atlantic picture and the Central/Eastern European picture that previous large aDNA Neolithic samples have involved with the latter involving more replacement and the former more assimilation).
I would have naively expected this kind of transition to be happening much earlier than 3000 BCE, which is thousands of years after the advent of the Neolithic in that region and is not all that earlier than the Bronze Age (i.e. right in the middle of the Copper Age and contemporaneous with high points of Sumerian society and relatively sophistication in Egypt relative to the rest of the world and prior to strong Semetic influence in Mesopotamia and contemporaneous with or a part of the tail end of the Atlantic European megalithic culture).
This suggests that common modern hgs which are very rare or absent in this sample (at least on the mtDNA side where there is more independence in descent) are Bronze Age or Iron Age arrivals. Comparing the U5 frequency then and now in the same area may also be a pretty useful way to roughly estimate the amount of population dilution from migration into the region that has taken place since 3000 BCE. This would suggest that something on the order 75%+ of the maternal gene pool in Southern France has a source outside Southern France within the last five thousand years, probably post-Copper Age rather than merely post-Paleolithic as previous ancient DNA studies have suggested.
The data I've seen on the timing of the European Neolithic would seem to fit the Neolithic in this part of France being a secondary expansion from an earlier Neolithic in Iberia, so affinity of hgs with Iberia doesn't tell us much about which population layer a hg comes from. Similarity could be a sign of an Cantabrian refugium source, but could just as easily also point to a source deriving from a first wave Iberian colony that is distinctive to the region due to founder effects.
The fact that we may have only a couple of founder males has some relevance to the possibility that this may explain the absence of J2/R1b, but given the homogenity of old populations in terms of hgs relative to modern populations, and the absence of ancient Y-DNA samples of these hgs in an ever expanding data set where there is independence, the case that men with hgs J2 or R1b were not nearly as common in the pre-Bronze Age era in Europe as they are now seems increasingly convincing.
"The G2a median joining network shows that the Treilles haplotypes are disjoint from those that dominate the North Caucasus,"
Looking closely at the median joining network, the case for the Northern Caucasus as the source of G2a almost anywhere else seems remote. Instead, the Northern Caucusus looks like a recipient of G2a from Mediterranean or the Middle East, and honestly, the Western Mediterranean looks like the best fit for a Northern Caucusus G2a source based on that network with the original G2a bearing men in the Northern Caucasus in this scenario probably arriving by sea, rather than overland from the Middle East, and the migration probably taking place at some time before R1b became common in the Mediterrean, but probably post-Neolithic revolution.
The network is also suggestive of the idea that the Trielles group may have had immediate antecedent in Italy and only more remove antecedents in Iberia.
This direction of migration is quite unexpected.
Among other things this would suggest that if there is a linguistic connection between the non-IE languages of Europe and the non-IE languages of the Northern Caucasus that the direction of linguistic evolution may have been from West to East rather than the other way around.
"Rather, there was a continuous process of migration over millennia".
And that seems to have been the case throughout the world. The Austronesian migration into the Pacific was far from being a simple affair. And I suspect the same is true of the entry into America and Australia. Even to the original expansion of H. sapiens (or even H. erectus) around the world.
"His ancestry is from the Irish Midlands, but has a grandmother from Kerry in the extreme South West of the country - who may account for the elevated Basque component".
Quite likely. Both Irish and Basques share a high proportion of R1b1a2a1a1b-P312. Obviously Y-chromosome is not the defining character but it probably shows some underlying connection.
The proof of the cake is in the eating. This I2a is clearly almost 3 times older than the younger Y-chromosome estimates. So the younger estimates are almost certainly wrong. This has implications across the Y-chromosome spectrum. Notable for R1b subclades.
I think we should stop talking about R1b as if R1b in western Europe is one phenomenon. It clearly isn't. There are at least two major groups that move differently and arguably more.
Iberian (U152/R1b1a2a1a1b3/R1b1b2a1a2);
Northsea (U106, R1b1a2a1a1a*, R1b1b2a1a1).
Everything I read fits with Indo European languages travelling latterly into Europe with an R1a haplogroup probably with West Asian/Balkan genes. It was probably the language of trade, like English, and dominated earlier languages because of this.
Argie where do you get your haplogroup statistics? They are higher than mine. I have been looking for a more recent haplogroup statistical database.
I was refering to an association with Semitic in the Near East not necessarily outside it.
Even in the Near East (I am having the Near East synonymous with West Asia) J2 is far from being associated with Semitic languages, as it is more concentrated in the non-Semitic speaking regions of the Near East such as Anatolia, the Armenian Highlands and the Transcaucasus than in the Semitic speaking regions of the Near East as a whole.
n/a: I have no idea where you get your draft from. Is there a paper supporting it or is this another FTNA-induced mess?
I wrote that entry on Myres 2010, which is a good and important paper on West European R1b. You cannot avoid that reference, it's not even one year old.
The "Southern haplogroup", in any case, has a scatter between Iberia and France with some extension, notably to the Atlantic islands quick impression: in blue). Only four non-private subclades are known: two are sub-Pyrenean (one exclusively Basque and the other more distributed), another is scattered through all Europe and yet another is scattered from South France to the Atlantic Islands.
Of the rest of R-S116, of the asterisk paragroup, it's all concentrated in Southern France and Iberia. So So I do not know if you have somehow detected a rare clade in, say, Denmark, but I know it will be present in SW Europe for sure, as are all other subclades of this linage, which is clearly of Franco-Cantabrian origin and also the largest sublineage of R1b.
"Of the currently categorized basal subclades of R-P312, none that I know of show evidence of origins in SW France or Iberia".
I did not say "SW France". The Franco Cantabrian region includes almost all France (but the North, which used to be poorly inhabited in the Paleolithic). But from the Loire southwards, it includes all. Notably, you cannot exclude SE France.
Per the current ISOGG tree R1b1a2a1a1b (S116) includes (non-private):
- R1b1a2a1a1b2-M153 (Basque-Gascon)
- R1b1a2a1a1b3-U152 (common in South France, sub-Pyrenean)
- R1b1a2a1a1b4-L21 (common in the Guyene and extending into the Atlantic islands)
- R1b1a2a1a1b5-L176.2 (including the old M167, clearly sub-Pyrenean)
- R1b1a2a1a1b6-L238 (of which you say: "I believe L238 is associated with Scandinavia" but is probably just poorly researched)
So, no matter how you twist it, the Franco-Cantabrian origins show up.
A problem with the data coming up from commercial sites (I hope they all succumb to crisis and regulations) is that almost all the data comes from NW Europe. These venues do not make balance samples and rely on the naivety or enthusiasm of participants, which typically come very particular areas that, as result, are oversampled, even if they were only colonized after the Ice Age (like Scandinavia or Britain).
The other side of the coin is those areas less interested in DNA testing, less fluent in English or simply less affluent, which are undersampled. In West Europe that means most of the interesting areas for this debate, notably France and Iberia.
So I'll wait for academic papers instead of relying on private companies. I must say here that I totally disagree with the renaming scheme applied to R1b on FTDNA arbitrariness. But well.
PS- "There's this thing called genetic drift".
Genetic drift only happens when populations are small and stable, not when they are large and expansive as since Neolithic. No relevant drift since Neolithic, please.
because your argument is that 3500 +/- 500 years ago a massive unique event, a wave of people flowed into Europe, making a specific genetic change in the y-dna, R1b1b2, whose diversity can be calculated so as to determine when that unique event happened.
Have you bothered to read anything in this blog before you claim that this is "my" argument. I have emphasized inte alia:
1. The huge confidence intervals associated with TMRCA estimates,
2. The interplay of population growth rate and the posterior probability of the TMRCA given the observed allele variance
3. The dependence of age estimates on the mix of slow/fast moving markers one uses.
Hence, I have made no strong claims about particular haplogroups on the basis of a supposed accurate TMRCA estimate, with the exception of E-V13 which not only appears to be one of the youngest major Y-haplogroups in Europe, but also has quite a lot of other evidence in favor of the association I claimed for it.
Europe wasn't a happy hunting ground where any foreigner could come over and set up shop and bring with him 1 million relatives. The only chance was 8000 years ago. After that, they became farmers, exploded in population and the window of opportunity was closed.
Your understanding of European demographic history is too simplistic. There were not only population explosions, but also population crashes. For example, the Tripolye mega-sites you refer to vanish from existence.
Nor is it true that the first farmers largely filled up the continent. The process as it is currently understood involves rapid leapfrog colonization, not slow and laborious population-driven demic diffusion.
Going back to the analogy of the Americas, after 500 years, its population density is still about an order of magnitude less than that of Europe, and the percentage of Native American ancestry in its population continues to diminish.
Or, the earliest Neolithic of the Near East took several thousand years before spilling into Europe. Apparently there was plenty of room there, so I don't really see how you can claim that the earliest European farmers immediately reached demographic saturation and there was no further migration into Europe since 8,000 years ago.
Argie where do you get your haplogroup statistics?
Annie, mostly from yhrd and ysearch results. But here are some results from studies:
Munich 8% G2a (Kayser, 2005)
North Italy 4% G2a (Di Giacomo, 2003)
Southeast France 8% G2a (King, 2011)
Iberia 5% G2a (Adams, 2008)
Note that the North Italy result was on the basis of 120 samples, but my estimate is on the basis of 1500 yhrd samples.
I believe L238 is associated with Scandinavia, and the little that I know of DF19 suggests it's more common in Northern Europe as well.
This is a few tested samples of a newly found SNP, right? That will reflect the hugely lopsided North European origin of FTDNA customers. In ysearch there are more SRY2627 samples from England than from France and Spain combined, but obviously SRY2627 is a lot more common in the latter 2.
Dienekes, why are all European people remarkably close to each other in FST distances? Bulgaria is much closer to England than to Turkey. Your point of view requires a total overhaul of the European population. All those massive waves from here and there would have produced a Disneyworld people with huge FST distances between each other, what with the Poles being Pakistanis with blond hair and the Swedes and Irish being Arabs with blue eyes, etc. And while we're at it, after this massive population overhaul just 3000 or 4000 years ago, how and why did North European Turks develop the most recessive trait known to mankind: blond hair and blue eyes? That's amazing, the Turks in England needed blue eyes to farm better??? But the Turks in Spain didn't?? And this all happened in 1500 years, just in time to be described as blond and blue eyed by the Romans???
Dienekes, how about making an admixture study of West Asians using only the X-chromosome, to see if there are any sex-based differences from the all-chromosomes results you posted at the Dodecad Project? This of course is in relation to the supposed expansions of Anatolians and other people in the last 3000 years.
Yeah. Population size increases and decreases.
Some older (half a century old) estaminates on German population development:
1th century: 1 Million
3th century: 3 Million
5th century: 1 Million (migration era. Germans heading to Britain, Frane and Italy
10th century: 4 Million
12th century: 7 Million
13th century: 12 Million
14th century: 6 Million (plague)
Today: roughly 80 Million
Not only do these numbers estaminate that Germany was a rather empty forrest only 2K years ago, but also, that the population size crashed several times.
One also needs to imagine Magyars raiding FRANCE. Remeber they need to cross the German kinddom (3 Million citicens at that time) to do so.
I imagine it was easy. The land was empty wilderness.
Dienekes said:
Speaking of the Basques, the most interesting result from the Dodecad Project is that they seem to lack the "West Asian" component than their French/Spanish neighbors possess. J2 was also not found in an ancient Basque cemetary.
pconroy replied:
As I commented recently my father (DOD098) has no known non-Irish ancestry, and also has NO "West Asian" component
Conroy, this is only true for Dienekes' alternative ADMIXTURE analysis:
http://dienekes.blogspot.com/2011/04/genetic-structure-of-west-eurasians.html
But in Dienekes' main ADMIXTURE analysis (the one Dienekes normally uses for the Dodecad Project) your father has 5.4 % "West Asian" component (and in it your father has even 1.4 % "South Asian" component). So clearly there is a difference between the "West Asian" components of the two ADMIXTURE analyses.
Basques, on the other hand, seem to lack the "West Asian" component entirely in both Dienekes' main ADMIXTURE analysis and Dienekes' above linked alternative ADMIXTURE analysis. So Basques seem to be less Indo-European admixed than your father, assuming that in both ADMIXTURE analyses the "West Asian" component in Western Europeans is totally or almost totally related to the spread of the Indo-European speakers.
Dienekes, why are all European people remarkably close to each other in FST distances? Bulgaria is much closer to England than to Turkey.
I don't know the Fst distances of Bulgarians to other populations, but Tuscans are genetically closer to Turks than to English according to the Fst tables I have (Greeks and South Italians must be genetically even closer to Turks and more distant to English than Tuscans are).
Maju,
Myres supports what I'm saying. The latest information, even more so. You can wait until it makes its way into print in a year or two, or you can pay attention now. I don't care which.
Note the acknowledgment in Myres: "We thank Vincent Vizachero for alerting us to the potential of M520, M529, S116, L11, L23 SNPs and for insight regarding the DYS390, 19 repeat allele and M73."
Vizachero, an amateur, pointed out the basic patterns visible in the Myres data long before that paper was published.
argiedude,
"This is a few tested samples of a newly found SNP, right? That will reflect the hugely lopsided North European origin of FTDNA customers."
L238 is associated with a previously identified "R1b Norse" STR signature.
DF19 was discovered using 1000 genomes project data, and has not yet been made available for testing at FTDNA. I believe many more of the L21- and U152- R-P312 1000 genomes samples are of Southern European origin (from Italy, Spain, and Latin America) than from Northern Europe, but so far DF19 has only been observed in two British samples and a CEU sample.
"Myres supports what I'm saying. The latest information, even more so".
Not at all. The best you have been able to muster so far is a lesser clade (probably detected in a oversampled Nordic state), with almost no known members which "you think" it has something to do with Sweden.
Vagueness! Wishful thinking! And does not even matter because it'd be 1 vs. 4 - plus all the brutal R1b1b2a1* still lying in hidden in like 80% of the people with R1b in SW Europe. So go and research SW Europe!
Fanty,
I agree that population size fluctuations are important, especially on a local scale (but less so on a continental scale). But I don't agree with "The land was empty wilderness." LBK quickly and decisively settled the best agricultural grounds: flat Loess soil valleys. Field rotation or heavy tilling were unknown, so after local resources were depleted, they moved on, but later cultures came back with a vengeance and for good. Steepish slopes could not be worked, and the band of nutrition-poor sandy soils from (now) the Netherlands and northern Germany to east of the Elbe river was not crossed for a millennium or more, and was not settled until population pressure forced people to live there (and very poorly at that until the recent advent of potato and corn farming and artificial fertilizers). But in between was dense original forest with impenetrable underbrush, not highways easily passed by anyone not invited. And no year-round or high-protein source of fodder for horses, by the way.
But yes, pockets of different populations likely means much, much lower effective population sizes than what is currently assumed. Something clearly is completely wrong with current time estimates. Differing mutation rates in different genetic regions are hard to blame, because mutations counts will always be dominated by the fastest ones - so the slow ones don't matter unless you really screw up your statistics. Which still is a slight possibility, of course.
At any rate, my view is that LBK was dominated by local Danubian mt- and y-DNA with some fraction of Anatolian mixed in, and picking up additional haplogroups along the way, increasingly so at the fringes. Then, after LBK collapse, the fringes (which in part contained DNA from the south and the southern Mediterranean route) started to dominate, leaving mostly pre-LGM haplotypes in Europe. Things did not even remotely come to some kind of mixed balance until about the bronze age. So, what we need is much larger numbers of local test sites and over different time periods: what we have now is not even remotely representative of an average description of haplogroups over time or space.
Dienekes, how about making an admixture study of West Asians using only the X-chromosome
Self reply: because that would result in just 10K SNPs, which is way less than the minimum needed to generate the West Asian clusters seen in the Dodecad Project. Still, it would be a good idea to do this X-chromosome-solo ADMIXTURE analysis to see about sex differences with respect to the bigger continental genetic clusters.
I don't know the Fst distances of Bulgarians to other populations, but
Good hijacking attempt. But back to my point, if it were true that R1b in West Europe is the result of something that happened 3500 years ago, then there should be a very noticeable genetic distance (FST) between West Europe (60% R1b) and East Europe (10% R1b). Not only is there no noticeable difference, but in fact the gradient of FST change is smaller in an east-west direction than in a south-north direction. In example, 1000 km north-south in Europe has an average FST of 0,0018, while 1000km east-west averages 0,0009 FST. And no, this is not because the south received black or North African ancestry, because the same ratio holds true in southeast Europe.
And though it may sound, I dunno, not serious perhaps, you can't escape the issue about the trivial little detail of the blond hair and blue eyes of North Europeans, a trivial meanignless characetristic that has big implications precisely because of its meaninglessness. The argument that most of West Europe was physically replaced by Anatolians 3500 years ago means the new people obviously didn't have all these typical charcateristics of modern North Europeans. They had to come about between 3500 ya and 2000 ya, when Romans first described these people. It sounds trivial... it's not. Keep in mind that American Indians retain a notable similarity to East Asians, despite 15000 to 30000 years separation.
, if it were true that R1b in West Europe is the result of something that happened 3500 years ago, then there should be a very noticeable genetic distance (FST) between West Europe (60% R1b) and East Europe (10% R1b). Not only is there no noticeable difference.
You are triply mistaken:
First, stop parading your "3500 years" strawman, as I never claimed R1b was 3500-years old in Europe.
Second, a sweep of Y-chromosomes by R1b in Western Europe need not necessarily have been associated with a huge shift in autosomal frequencies. It only takes a few generations of mating with locals for an autosomal component to be diluted; moreover, the dilution would increase away from the point-of-origin of R1b
Third, there is indeed evidence for a relationship between R1b-rich populations and West Asia compared to R1a-rich populations. For example at K=4 in my recent analysis of West Eurasians:
http://dienekes.blogspot.com/2011/04/genetic-structure-of-west-eurasians.html, the West Asian component reaches much higher prevalence in Western Europe than in Eastern Europe. It is 13% in Irish vs. 1.3% in HGDP Russians and >30% in Iberians, and >50% in C/S Italians/Greeks and >60% in populations from West Asia.
So, I consider it quite likely that an R1b-rich population may have carried with it a "West Asian" autosomal component that was gradually diluted as it spread from West Asia to the Atlantic.
"... the West Asian component reaches much higher prevalence in Western Europe than in Eastern Europe"...
Not sure at what K-depth you mean but at K=11 Irish and Russians show similar "West Asian" component - quite low in both cases.
What is interesting in that analysis is that the West Asian components ("West Asian" and "SW Asian") only show up strong in Italy and the Balcans (Romania) in fact.
On the other hand there are a number of European populations that show NO or almost no presence of West Asian specific blood: Finnish, Lithuanians, Orcadians, Norwegians, Swedes, Sardinians (rather unexpectedly) and Basques.
"So, I consider it quite likely that an R1b-rich population may have carried with it a "West Asian" autosomal component that was gradually diluted as it spread from West Asia to the Atlantic".
Homeopathic type of dilution it seems to me because some of the strongest populations in R1b, like Basques, have less West Asian blood than active component is in an homeopathic remedy. Instead populations with relatively high levels of West Asian blood, have low levels of R1b (Italians, Balcanic peoples).
I'm really sorry but I'm more likely to believe in sugar pills.
Not sure at what K-depth you mean but at K=11 Irish and Russians show similar "West Asian" component - quite low in both cases.
I already said "K=4", which is also the first one that distinguishes between European and non-European Caucasoids.
K=11 is irrelevant for the issue at hand, as the components at K=11 represent regional variation, and not the major West Asian/European cline that dominates Caucasoid genetic variation.
On the other hand there are a number of European populations that show NO or almost no presence of West Asian specific blood: Finnish, Lithuanians, Orcadians, Norwegians, Swedes, Sardinians (rather unexpectedly) and Basques.
First, see above.
Second, even 0% "West Asian" does NOT imply an absence of "West Asian" admixture, it only implies that this admixture is minimum over the set of studied populations.
See:
http://dienekes.blogspot.com/2011/03/note-of-caution-on-admixture-estimates.html
Good hijacking attempt.
I did not attempt to hijack the thread, I just presented a broader context for the Fst issue.
"I already said "K=4""
My bad, I did not see that.
I will assume then that it is the "W/S Asian" component (middle blue).
I'm not sure I can accept these shallow depts, when deeper ones contradict them, i.e. any "West Asian" component vanishes in many of the studied populations. This is the same kind of problem that afflicts 1990s Cavalli-Sforza's PC1 and even as recently as 2007 at the low components of Bauchet's paper (but not at the deeper ones). The problem is that low K components tend to vanish in many populations as their real uniqueness is unveiled at deeper Ks.
We get to assume (because the program works that way) that West/South Eurasians are divided in two, and to some extent that is true. But it is a very shallow extent and ultimately meaningless. I'd be nice if these structure algorithms would include a default category for "neither this nor that" because then I am sure many populations that are now split in two at K=2 (K=4 here), etc. will no doubt have that "no affinity" pseudo-cluster as their main home, instead of being forced here or there.
Any mathematician with the skills to develop this so much needed algorithm, with an "other" option?
"... the components at K=11 represent regional variation, and not the major West Asian/European cline that dominates Caucasoid genetic variation".
I would question that to at least some extent. I think that that you have done a great job spotting the real multicolor variation of West Eurasians, which in this particular experiment is best captured at K=10 or lower. The differences between Turks and Germans (specially if you believe in Neolithic replacement) are "regional variation" (i.e. distinct from the great continental differences we sometimes call "races" - much more diluted and hard to spot). For that reason I think that assuming that the first dual clustering that the algorithm is able to form is something definitive implies a strong assumption.
"Second, even 0% "West Asian" does NOT imply an absence of "West Asian" admixture, it only implies that this admixture is minimum over the set of studied populations".
Same thing in practical terms. How can it be then that at K=4, say, Basques show some 20% W/S Asian component and at K=10 it is zero (minimum). There's a huge difference between zero (minimum) and 20% and that's why I trust more the K level that is able to capture what is Basque-specific (an specificity that has been confirmed once and again, not any accident).
I have been puzzling over England and Ireland. They are almost autosomally identical. The have essentially identical mitochondrial haplogroups, but dramatically different R1b patterns. Ireland is overwhelmingly dominated by Iberian R1b1a2a1a1b (79% R1b). England has a very substantial North Sea R1b1a2a1a1a (about half the R1b)as well as R1b1a2a1a1b (67% R1b).
These major differences in Y-haplogroup seem to have made almost NO difference to the autosomal DNA. This is very difficult to understand. Either
(1) These incoming R1b haplogroups rose slowly to dominance from a small base (therefore no autosomal change). Perhaps there was a breeding advantage (patriarchy maybe).
(2) The populations that these R1b haplogroups came from were ethnically identical to the Isle populations they overwhelmed.
(3) These haplogroups arose in situ from within a homogeneous ethnic group.
Ireland is prone to cycles of population bust and boom. So it was also possible that the dominance of Iberian R1b is due a reexpansion from a small base, but then I would expect that to affect the mitochondrial haplogroups too. So this does not seem to be the cause.
R1b haplogroups in England and Ireland really do NOT look like recent mass migrations of ethnically different men from the east of Europe. Especially not two separate migrations of men in recent times. One from south the other from the north/center.
IMO it looks like the people of the Isles are a homogeneous people (North and South blended somewhere else). My working hypothesis is that the difference in Y haplogroups is due to local patriarchy after arrival in the Isles. Ireland was a separate area and subject to more economic upheaval than England. Hence a more extreme effect.
The R1b haplogroups could be indigenous to the population or introduced with the patriarchy. What is clear is that they rose to dominance WITHIN the ethnicity, not as an sudden external takeover, as the basic autosomal ethnicity has not been disturbed.
The rest of Europe is rather more complicated. It certainly looks like R1b arose in the east, travelled north of the med. But when?
"Everything I read fits with Indo European languages travelling latterly into Europe with an R1a haplogroup probably with West Asian/Balkan genes. It was probably the language of trade, like English, and dominated earlier languages because of this".
That would be my interpretation of the situation also.
"I consider it quite likely that an R1b-rich population may have carried with it a "West Asian" autosomal component that was gradually diluted as it spread from West Asia to the Atlantic".
Yes. Most seem to believe that haplogroup expansion is always automatically associated with autosomal DNA expansion.
"Homeopathic type of dilution it seems to me because some of the strongest populations in R1b, like Basques, have less West Asian blood than active component is in an homeopathic remedy".
A 'private' lineage that expanded once it reached Western Europe? That is the explanation you've used to explain a similar problem with mtDNA in SE Asia. Of course you're aware of my belief that such an explanation is very unlikely.
http://anpron.eu/wp-content/uploads/2011/06/Ancient-DNA-reveals-male-diffusion-through-the-Neolithic-Mediterranean-route.pdf
"Ireland is overwhelmingly dominated by Iberian R1b1a2a1a1b (79% R1b). England has a very substantial North Sea R1b1a2a1a1a (about half the R1b)as well as R1b1a2a1a1b (67% R1b)."
England bore the brunt of Norman and Viking invasions in the Middle Ages. Ireland didn't. The North Sea hg of R1b is a fit to this scenario.
England bore the brunt of Norman and Viking invasions in the Middle Ages. Ireland didn't. The North Sea hg of R1b is a fit to this scenario.
Not true. The Vikings and the Normans had a significant presence in Ireland. In fact, many common Irish surnames such as Doyle, Fitzgerald and Burke indicate Viking or Norman ancestry. Also, if North Sea R1b were brought to Britain by the Vikings and the Normans then we should also see high levels of R1a1 as well which we don't. I think the differences in R1b types in Ireland compared to Britain is due to a "Niall of the Nine Hostages"-effect where the highest status Irishmen were producing the vast majority of the children. For whatever reasons - probably luck - Iberian R1b males gained dominance over North Sea R1b males in Ireland and were, therefore, able to outbreed them.
Ireland is overwhelmingly dominated by Iberian R1b1a2a1a1b (79% R1b). England has a very substantial North Sea R1b1a2a1a1a (about half the R1b)as well as R1b1a2a1a1b (67% R1b).
Could it be possible that the high levels of North Sea R1b in Britain are due to the Anglo-Saxon invaders? After all, it would explain why the English became a Germanic-speaking people while the Irish remained Celtic-speaking.
"Ireland is overwhelmingly dominated by Iberian R1b1a2a1a1b (79% R1b)"
R-P312 is not "Iberian". R1b expanded into Ireland, England, and Iberia via central Europe.
There are other plausible explanations, Joe, notably the Maglemosean culture (Epipaleolithic, rooted in Ahrensburgian AFAIK), which extended at both sides of the North Sea, being probably originally based at least partly on the then sinking Doggerland. I am not sure of all the details but I understand that while south and west Britain (and hence surely Ireland) was colonized from France across the Channel, NE Britain was surely colonized by people related to those in the Netherlands, Low Germany and Denmark.
Another possible moment of differential colonization is the Neolithic, when most of England was colonized (?) by peoples of modified Danubian culture from NW France, while Cornwall, Wales and Scotland (and hence maybe Ireland) were from the Megalithic zone of Brittany and High Normandy.
And then of course the Anglo-Saxons and Danes (Vikings) may have reinforced that pre-existent pattern - but only reinforced. We should not expect that the impact of Anglo-Saxons/Danes would be greater than the apportion of R1a (for example) suggests. So, if R1a is 16% in Denmark and only 6% in England (data taken from Wikipedia), then the impact of Germanic flows cannot be greater than 6/16=0.375 or 38%.
That's still pretty big but cannot explain the 57% of R1b1a2a1a1a (R-U106), compared with only 35% in Denmark (because 35x0.38=13, so only 13 percentual points of that 57% English R1b1a2a1a1a can be attributed to Germanic invaders. The rest must be older.
Denmark is probably not the optimal reference point as its physical connection to the continent probably was less of a barrier to subsequent gene flow than on the other side of the Baltic Sea.
At any rate, Maju's back of napkin estimate suggests that the R1a v. R1b (North Sea hg) ratio is consistent with Vikings as the single largest source of both hgs in Britain.
The point is that the autosomal profile is the same (near as)for England and Ireland. So any differential incoming haplogroups did not disturb the autosomal base. If is Viking or Anglosaxon sourced then these population whould have to be genetically identical (not likely for the Vikings, possible for Anglosaxons). But as I recall the specific subgroups previously ruled out significant Anglo Saxon input.
Don't be nitty-picky, Andrew: it's for me a lot easier to find data for Denmark than for Swchelswig-Holstein or Low Saxony (as separate entities within Germany). It is known that their genetics are similar and I cannot say the same re. all-German genetics. Low Germany is a distinct genetic region that approaches better Denmark than High Germany or even East Germany.
"At any rate, Maju's back of napkin estimate suggests that the R1a v. R1b (North Sea hg) ratio is consistent with Vikings as the single largest source of both hgs in Britain".
That's not what I gather: I assumed that at most 100% of R1a is of Germanic origin. Part fo it could have arrived with earlier IE flows like Celts from further South but I do not feel like making such complex estimates and I'm hence assuming that 100% of arrived with Germanics (not just Vikings, actually mostly Anglos, Saxons and Jutes). But this is a maximum figure in any case.
Based on that I gathered that at most 13 percentual points out of 57 total of R1b1a2a1a1a (R-U106) should have arrived from that same Medieval continental source. That is 23% of the total R1b1a2a1a1a (R-U106) and not "the single largest source". The rest should be older and hence have other source(s), as considered above.
"R-P312 is not 'Iberian'. R1b expanded into Ireland, England, and Iberia via central Europe".
Perhaps it's not 'Iberian', but R1b1a2a1a1b1b4-L21 cannot have coalesce in Ireland before the Holocene. That is it can be no older than about 10,000 years. Is the whole R1b1a2a1a1b1b-P312 expansion about the same age? Perhaps. Did that expansion begin in SW Europe? Perhaps. But R1b1a2a1a1b1b-P312 shares a region of origin with R1b1a2a1a1b1a-U106, which is basically northern, probably North European Plain. Did the two haplogroups survive as remnants of the Gravettian? Perhaps. Where is R1b1a2a1b-L277 found? That might give us a clue. To me it looks very much as though R1b1a-P297 had spread itself from Anatolia to Afghanistan long ago. About the time that R1b1c-V88 made it to Afria, whenever that was.
I don't know the Fst distances of Bulgarians to other populations, but Tuscans are genetically closer to Turks than to English according to the Fst tables I have (Greeks and South Italians must be genetically even closer to Turks and more distant to English than Tuscans are).
I argued this on the blog a few months back.
I have had extensive personal contact with these three ethnic groups in London and it seems obvious to the eye that Greeks are genetically much closer to Turks than to the English. They are also more similar culturally, in food and music for instance.
Quick question to the group: how certain are you that the bones in question cover the 200-year span of time? The best info on the age of the bones in the Treilles Cave I was able to find was a "Probable date" of 3,500-2,000 BCE (Maryvonne Naudet and Raymond Vidal, don't know the pub date). As far as I can tell, this comes directly from reference source #14 from the PNAS paper. It appears to be a monograph (in French) and I can't find it.
This article and the comments to it cover many subjects of interest to me. I have recently been investigating haplogroup G, so it is interesting to find some more ancient examples of it. Unfortunately G2a is a very diverse group with many branches, some found mainly in the Middle East (especially the Caucasus), some mainly in Europe, and some in both regions. Therefore it is a pity that we do not know which of them is represented here. The STRs given are not particularly good for identifying the branches. DYS392 = 11 shows that it is not G2a1, since that group has 392 = 10. Most of the examples in Ysearch belong to G2a3, characterized especially by DYS425 = 14. G2a3 also has many sub-divisions, extending from the Middle East to Europe. Its most important branch in Europe is G2a3b1a2, characterized by DYS388 = 13 and DYS617 = 12. This makes up a very large part of haplogroup G in Western Europe, but it rarely occurs east of Germany and Italy. I think its position within G2a3 is rather similar to the position of R1b1a2a1a1 L11 within R1b1a2 M269.
The subject of R1b1a2, its branches, their relationship to autosomal DNA is not very closely connected with the original subject, but it has attracted considerable attention in the comments. Since it is also of great interest to me, I will add my contribution. I think it is a mistake to describe R1b1a2a1a1-b S116 as “Iberian”. This group is common across the whole of Western and Central Europe, and it forms the majority of R1b almost everywhere. The big exception is the Netherlands, North Germany and Denmark, where R1b1a2a1a1-a U106 predominates. R1b1a2a1a1-b2 M153 is common in South-West Europe, but rare in the British Isles. I think this is evidence against the idea of a special connection between the Isles and SW Europe.
The R1b1a2a1a1 in Ireland is mainly R1b1a2a1a1-b4 L21. This group is widespread on the continent, but with much lower percentages than in Ireland. In addition, Irish R1b1a2a1a1-b4 L21 is largely made up of specific sub-branches. The most important of these is R1b1a2a1a1-b4b M222 characterized by DYS392 = 14 and DYS448 = 18. This is the group associated with Niall of the Nine Hostages. There are others, for example, one characterized by DYS617 = 13, 572 = 12, 640 = 12, and another with YCA IIb = 24 and DYS531 = 12. These groups are common in Ireland and Scotland, rare in England and almost absent on the continent. I think research should be done to find out if R1b1a2a1a1-b4 from Ireland has a close connection with R1b1a2a1a1-b4 in any particular part of the continent.
In England, R1b1a2a1a1-a U106 and R1b1a2a1a1-b3 U152 are more numerous and there is an increased presence of haplogroups other than R1b. It seems that R1b1a2a1a1-b4 L21 must have come to the Isles early and developed distinct branches there. The groups found more in England presumably came later. The Anglo-Saxons and Danes must have made a significant contribution, but it is possible that the difference between y-chromosomes in England and Ireland originated even earlier. I need to collect more evidence on this. I have also looked at the autosomal evidence, especially in Dodecad to see if there are any differences between Ireland and England that may be related to the differences in y-haplogroups. As several people have written above, such differences in autosomal DNA have not appeared so far. It is possible that they do not exist, but I have thought of another possibility. The Dodecad analyses include people from the whole world. Perhaps differences between people from different parts of the British Isles and North-West Europe are too small to appear when comparisons are made with Africans and Asians. I suggest that people, who are as purely Irish or purely English as possible should be compared with each other and with people from the nearest part of the continent. Perhaps this would reveal some interesting differences. The differences between the different branches of R1b1a2a1a1 are actually very small in the context of the overall diversity of the human y-chromosome.
Thanks Styan. That is exactly the sort of information I've been looking for.
The Dodecad and Eurogenes analyses can pick up autosomal differences between English/Irish and other nearby western European nations. Even between Scandinavians. I think even Portugal sticks out from Spain and the Basques certainly do. England does have a few extra very minor differences from Ireland (eg African) but the overall very deep similarity to Ireland is unaffected. Certainly there is nothing that is of the scale of the Y-haplogroup differences.
You can also look at individual profiles of Irishfolk and English folk at Eurogenes and Dodecad and there ARE stark differences. But they are not between the English and the Irish. I think there will be county/shire differences, with rural places like Galway and parts of northern England retaining more basic structure than coastal towns/cities. But these iron out to almost identical.
I am a g2a whose paternal line goes back to France. I just got my autosomal results, and I have a question for you. It lists me as 91.5 percent Western Europe, French and Orcadian. This makes sense to me because this is where my documented origins go. It also said I am 8.5 percent Middle Eastern (Palestinian/Jewish/Mozabite/Bedouin/Druze)
I imagine I could be part Jewish, but the rest does not fit any modern documentation, and they say that this is supposed to be about recent ancestry. I've got almost all of my lines 12-13 generations back to France and the British Isles. Is this perhaps a reflection of Neolithic farmers? Has anyone determined how common Middle Eastern genes are in Western European autosomal testing?
Thanks
Post a Comment