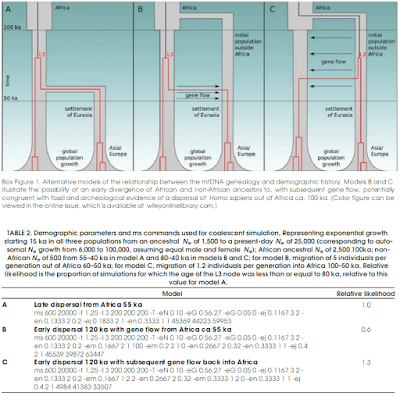
An excellent review which -among its other graces- demolishes the view that mtDNA haplogroup L3 provides a terminus post quem of 70 thousand years for the Out-of-Africa expansion, a question I've discussed in this blog before.
I think the evidence is overwhelming at this point that there were modern humans outside Africa before 100,000 years ago. The argument that they were a failed expansion is shoddy and is based, as far as I can tell on things like the age of L3, the assumption that Y-chromosome haplogroup E is native to Africa and not derived from back-to-Africa migrants, the assumption that Out-of-Africa coincided with the Upper Paleolithic cultural efflorescence (disproven by the earlier dating of Neandertal admixture), or the failed hypothesis of a coastal route Out of Africa 60 thousand years ago that seems to be repeated in inverse proportion to the evidence for it. The halving of the human autosomal mutation rate relative to what was inferred before has certainly not helped either.
Rethinking the dispersal of Homo sapiens out of Africa
Huw S. Groucutt, Michael D. Petraglia, Geoff Bailey, Eleanor M. L. Scerri, Ash Parton, Laine Clark-Balzan, Richard P. Jennings, Laura Lewis, James Blinkhorn, Nick A. Drake, Paul S. Breeze, Robyn H. Inglis, Maud H. Devès, Matthew Meredith-Williams, Nicole Boivin, Mark G. Thomas andAylwyn Scally
Current fossil, genetic, and archeological data indicate that Homo sapiens originated in Africa in the late Middle Pleistocene. By the end of the Late Pleistocene, our species was distributed across every continent except Antarctica, setting the foundations for the subsequent demographic and cultural changes of the Holocene. The intervening processes remain intensely debated and a key theme in hominin evolutionary studies. We review archeological, fossil, environmental, and genetic data to evaluate the current state of knowledge on the dispersal of Homo sapiens out of Africa. The emerging picture of the dispersal process suggests dynamic behavioral variability, complex interactions between populations, and an intricate genetic and cultural legacy. This evolutionary and historical complexity challenges simple narratives and suggests that hybrid models and the testing of explicit hypotheses are required to understand the expansion of Homo sapiens into Eurasia.
Link and here









