A very interesting paper that addresses the question of whether a selective sweep proceeds from standing variation (i.e., an allele already exists in the population, perhaps for a long time, and becomes "advantageous" only when it is paired with the right environmental stimulus), or from a new mutation (i.e., the selection pressure begins first, and a new allele appears by mutation and gets positively selected).
This question is of interest to me, because it might help interpret the occurrence of alleles that may be selected in one core region -where, perhaps, the selection pressure is highest, or they've had the most time to increase in frequency- but also occur at low or even trace frequencies in many more regions.
If selection occurs from standing neutral variation, then the occurrence of the allele in a wide geographical region is not particulary noteworthy; presumably the allele occurred at such frequencies in many places, but became selected in a few.
On the other hand, if an allele occurs from de novo mutation, then it's low frequency occurrence outside its core region is evidence of gene flow, and perhaps recent one. This gene flow may be facilitated by the selection pressure itself (i.e., when people move with the technology, e.g., milk, that creates this pressure in the first place).
PLoS Genet 8(10): e1003011. doi:10.1371/journal.pgen.1003011
Distinguishing between Selective Sweeps from Standing Variation and from a De Novo Mutation
Benjamin M. Peter et al.
An outstanding question in human genetics has been the degree to which adaptation occurs from standing genetic variation or from de novo mutations. Here, we combine several common statistics used to detect selection in an Approximate Bayesian Computation (ABC) framework, with the goal of discriminating between models of selection and providing estimates of the age of selected alleles and the selection coefficients acting on them. We use simulations to assess the power and accuracy of our method and apply it to seven of the strongest sweeps currently known in humans. We identify two genes, ASPM and PSCA, that are most likely affected by selection on standing variation; and we find three genes, ADH1B, LCT, and EDAR, in which the adaptive alleles seem to have swept from a new mutation. We also confirm evidence of selection for one further gene, TRPV6. In one gene, G6PD, neither neutral models nor models of selective sweeps fit the data, presumably because this locus has been subject to balancing selection.
Link
Showing posts with label ASPM. Show all posts
Showing posts with label ASPM. Show all posts
December 15, 2012
November 26, 2008
Allele surfing vs. Positive Selection
Continental human populations have very high allele frequency differences in several loci. One explanation for this phenomenon is that after their arrival in new lands, humans underwent selection for alleles that were appropriate in the new environments. An alternative explanation is that the frequencies were due to allele surfing, a process in which a small subset of individuals at the frontier of the expansion expands and multiplies into previously unsettled territory, causing their particular alleles to increase in frequency there.
From the paper:
This is a serious challenge to the selectionist paradigm and should be answered by its proponents. I would say that, from now on, the "gold standard" of positive selection should be concrete evidence that the proposed selected alleles actually do something that could have been selected, e.g., lactase persistence, where allele frequency differences are combined with a specific trait, which in turn is correlated with a particular selective influence (milk consumption after weaning). Statistical inference of selection without a comprehensive explanation is no longer intellectually convincing.
For example, ASPM and MCPH1 are loci that generated a lot of excitement as selection targets due to their large inter-group frequency differences. However, followup work has not found any substantial associations between them and anything of value: Has ASPM been the target of recent selection?, ASPM, MCPH1, CDK5RAP and BRCA1 and general cognition, reading or language. Were they really selected, or did they ride the wave of human advance?
Annals of Human Genetics doi: 10.1111/j.1469-1809.2008.00489.x
Large Allele Frequency Differences between Human Continental Groups are more Likely to have Occurred by Drift During range Expansions than by Selection
T. Hofer et al.
Several studies have found strikingly different allele frequencies between continents. This has been mainly interpreted as being due to local adaptation. However, demographic factors can generate similar patterns. Namely, allelic surfing during a population range expansion may increase the frequency of alleles in newly colonised areas. In this study, we examined 772 STRs, 210 diallelic indels, and 2834 SNPs typed in 53 human populations worldwide under the HGDP-CEPH Diversity Panel to determine to which extent allele frequency differs among four regions (Africa, Eurasia, East Asia, and America). We find that large allele frequency differences between continents are surprisingly common, and that Africa and America show the largest number of loci with extreme frequency differences. Moreover, more STR alleles have increased rather than decreased in frequency outside Africa, as expected under allelic surfing. Finally, there is no relationship between the extent of allele frequency differences and proximity to genes, as would be expected under selection. We therefore conclude that most of the observed large allele frequency differences between continents result from demography rather than from positive selection.
Link
From the paper:
The survey of the HGDP database on human polymorphisms reveals that large allele frequency differences between continental regions are extremely common. Indeed as much as 30% of loci show very large allele frequency differences between continents. These differences are unlikely to have been created by positive selection, but are more likely the result of neutral demographic processes such as the surfing phenomenon. Because the erosion of large allele frequency differences by mutation is slow, even for large mutation rates, the surprisingly large number of strongly differentiated STR alleles also do not need to be explained by the action of positive selection. Africa and the Americas show a much larger extent of differentiation than Eurasia or East Asia, which is certainly due to changes in allele frequencies during the colonisation of the Eurasian and the American continents. Disentangling the effects of selection and neutral demographic processes on genome diversity remains an important challenge of future human evolution studies.
This is a serious challenge to the selectionist paradigm and should be answered by its proponents. I would say that, from now on, the "gold standard" of positive selection should be concrete evidence that the proposed selected alleles actually do something that could have been selected, e.g., lactase persistence, where allele frequency differences are combined with a specific trait, which in turn is correlated with a particular selective influence (milk consumption after weaning). Statistical inference of selection without a comprehensive explanation is no longer intellectually convincing.
For example, ASPM and MCPH1 are loci that generated a lot of excitement as selection targets due to their large inter-group frequency differences. However, followup work has not found any substantial associations between them and anything of value: Has ASPM been the target of recent selection?, ASPM, MCPH1, CDK5RAP and BRCA1 and general cognition, reading or language. Were they really selected, or did they ride the wave of human advance?
Annals of Human Genetics doi: 10.1111/j.1469-1809.2008.00489.x
Large Allele Frequency Differences between Human Continental Groups are more Likely to have Occurred by Drift During range Expansions than by Selection
T. Hofer et al.
Several studies have found strikingly different allele frequencies between continents. This has been mainly interpreted as being due to local adaptation. However, demographic factors can generate similar patterns. Namely, allelic surfing during a population range expansion may increase the frequency of alleles in newly colonised areas. In this study, we examined 772 STRs, 210 diallelic indels, and 2834 SNPs typed in 53 human populations worldwide under the HGDP-CEPH Diversity Panel to determine to which extent allele frequency differs among four regions (Africa, Eurasia, East Asia, and America). We find that large allele frequency differences between continents are surprisingly common, and that Africa and America show the largest number of loci with extreme frequency differences. Moreover, more STR alleles have increased rather than decreased in frequency outside Africa, as expected under allelic surfing. Finally, there is no relationship between the extent of allele frequency differences and proximity to genes, as would be expected under selection. We therefore conclude that most of the observed large allele frequency differences between continents result from demography rather than from positive selection.
Link
September 25, 2008
Varki et al. (2008) on Human Uniqueness in Nature Reviews Genetics
From the paper:
Nature Reviews Genetics doi:10.1038/nrg2428
Human uniqueness: genome interactions with environment, behaviour and culture
Ajit Varki et al.
Abstract
What makes us human? Specialists in each discipline respond through the lens of their own expertise. In fact, 'anthropogeny' (explaining the origin of humans) requires a transdisciplinary approach that eschews such barriers. Here we take a genomic and genetic perspective towards molecular variation, explore systems analysis of gene expression and discuss an organ-systems approach. Rejecting any 'genes versus environment' dichotomy, we then consider genome interactions with environment, behaviour and culture, finally speculating that aspects of human uniqueness arose because of a primate evolutionary trend towards increasing and irreversible dependence on learned behaviours and culture — perhaps relaxing allowable thresholds for large-scale genomic diversity.
Link
Remarkable similarities of known human and chimpanzee protein sequences initially led to the suggestion that significant differences might be primarily in gene and protein expression, rather than protein structure6. Further analysis of alignable non-coding sequences affirmed this ~1% difference. However, the subsequent identification of non-alignable sequences that were due to small- and large-scale segmental deletions and duplications21–23 showed that the overall difference between the two genomes is actually ~4%.
...
Why are coding-sequence changes in brain genes under a larger degree of purifying selection than in other tissues? The reason for this is not immediately clear as a wide range of brain function supports life to reproductive age in humans.
...
But this notion, which is based on single nucleotide changes in protein-coding sequence, has to be reconciled with the CNV data, because CNVs in humans seem to be enriched among genes involved in neurodevelopmental processes.
...
However, connecting such genes involved in disorders of human cognition to the specific phenotypes undergoing selection poses significant challenges. A salient example involves two genes, abnormal spindle homologue microcephaly associated (ASPM) and microcephalin (MCPH1), the adaptive evolution of these genes in humans was claimed to be related to normal variation in brain size, on the basis of the fact that Mendelian mutations in each results in microcephaly in humans152,153. However, not all investigators have found evidence for the adaptive evolution of ASPM or MCPH1 (ref. 154). Also, neither gene is likely to contribute significantly
to normal variation in human brain size155. This case illustrates the challenges of interpreting genetic data in the face of complex phenotypes, especially those that are poorly understood.
Nature Reviews Genetics doi:10.1038/nrg2428
Human uniqueness: genome interactions with environment, behaviour and culture
Ajit Varki et al.
Abstract
What makes us human? Specialists in each discipline respond through the lens of their own expertise. In fact, 'anthropogeny' (explaining the origin of humans) requires a transdisciplinary approach that eschews such barriers. Here we take a genomic and genetic perspective towards molecular variation, explore systems analysis of gene expression and discuss an organ-systems approach. Rejecting any 'genes versus environment' dichotomy, we then consider genome interactions with environment, behaviour and culture, finally speculating that aspects of human uniqueness arose because of a primate evolutionary trend towards increasing and irreversible dependence on learned behaviours and culture — perhaps relaxing allowable thresholds for large-scale genomic diversity.
Link
August 22, 2008
ASPM and cerebral cortex evolution in primates
ASPM was initially identified in human-chimp comparisons. Since it was expressed in the brain, it was thought that it played a role in making us different from our closest relatives. This new paper shows that adaptive evolution of ASPM is not limited in the human-chimp split, but occurred in many different primate lineages. Moreover, the target of its evolution was the cerebral cortex.
Note that these results are not directly applicable to the recently selected variant within the human lineage (some recent discussion).
Molecular Biology and Evolution, doi:10.1093/molbev/msn184
Positive selection in ASPM is correlated with cerebral cortex evolution across primates but not with whole brain size
Farhan Ali, and Rudolf Meier
The rapid increase of brain size is a key event in human evolution. ASPM (abnormal spindle-like microcephaly associated) is discussed as a major candidate gene for explaining the exceptionally large brain in humans but ASPM’s role remains controversial. Here we use codon-specific models and a comparative approach to test this candidate gene that was initially identified in Homo-chimp comparisons. We demonstrate that accelerated evolution of ASPM ( = 4.7) at 16 amino acid sites occurred in nine primate lineages with major changes in relative cerebral cortex size. However, ASPM’s evolution is not correlated with major changes in relative whole brain or cerebellum sizes. Our results suggest that a single candidate gene such as ASPM can influence a specific component of the brain across large clades through changes in a few amino acid sites. We furthermore illustrate the power of using continuous phenotypic variability across primates to rigorously test candidate genes that have been implicated in the evolution of key human traits.
= 4.7) at 16 amino acid sites occurred in nine primate lineages with major changes in relative cerebral cortex size. However, ASPM’s evolution is not correlated with major changes in relative whole brain or cerebellum sizes. Our results suggest that a single candidate gene such as ASPM can influence a specific component of the brain across large clades through changes in a few amino acid sites. We furthermore illustrate the power of using continuous phenotypic variability across primates to rigorously test candidate genes that have been implicated in the evolution of key human traits.
Link
Note that these results are not directly applicable to the recently selected variant within the human lineage (some recent discussion).
Molecular Biology and Evolution, doi:10.1093/molbev/msn184
Positive selection in ASPM is correlated with cerebral cortex evolution across primates but not with whole brain size
Farhan Ali, and Rudolf Meier
The rapid increase of brain size is a key event in human evolution. ASPM (abnormal spindle-like microcephaly associated) is discussed as a major candidate gene for explaining the exceptionally large brain in humans but ASPM’s role remains controversial. Here we use codon-specific models and a comparative approach to test this candidate gene that was initially identified in Homo-chimp comparisons. We demonstrate that accelerated evolution of ASPM (
 = 4.7) at 16 amino acid sites occurred in nine primate lineages with major changes in relative cerebral cortex size. However, ASPM’s evolution is not correlated with major changes in relative whole brain or cerebellum sizes. Our results suggest that a single candidate gene such as ASPM can influence a specific component of the brain across large clades through changes in a few amino acid sites. We furthermore illustrate the power of using continuous phenotypic variability across primates to rigorously test candidate genes that have been implicated in the evolution of key human traits.
= 4.7) at 16 amino acid sites occurred in nine primate lineages with major changes in relative cerebral cortex size. However, ASPM’s evolution is not correlated with major changes in relative whole brain or cerebellum sizes. Our results suggest that a single candidate gene such as ASPM can influence a specific component of the brain across large clades through changes in a few amino acid sites. We furthermore illustrate the power of using continuous phenotypic variability across primates to rigorously test candidate genes that have been implicated in the evolution of key human traits.Link
June 06, 2008
ASPM, MCPH1, CDK5RAP and BRCA1 and general cognition, reading or language
See also:
Intelligence doi:10.1016/j.intell.2008.04.001
Recently-derived variants of brain-size genes ASPM, MCPH1, CDK5RAP and BRCA1 not associated with general cognition, reading or language
Timothy C. Bates et al.
Abstract
Derived changes in genes associated with primary microcephaly (MCPH) have been suggested to be “currently sweeping to fixation” i.e., increasing in frequency in most populations, with the likely outcome that the derived allele will completely displace the ancestral allele over time. Possible causes for this sweep include effects on human reasoning and language. Here we test the hypothesis that these derived alleles are associated with current variation in spoken or written language and related traits. The association of derived alleles of the ASPM, MCPH1, CDK5RAP2 and BRCA1 genes was tested against well-validated measures of dyslexia, specific language impairment, working memory, IQ, and head-size in a family-based association study of over 1776 subjects from 789 families of twins. No evidence for association was found for any gene to any trait. The results strongly did not support the hypothesis that derived alleles in MCPH-related genes are related to the evolution of human language or cognition. Results were compatible with the alternate hypothesis, suggesting that adaptations in these genes associated with a dramatic increase in brain size have long since reached fixation and are now maintained by stabilizing selection.
Link
- Microcephalin and ASPM do not Account for Brain Size Variability
- ASPM and the alphabet
- Has ASPM been the target of recent selection? (*)
- ASPM and Microcephalin don't make people smarter
Intelligence doi:10.1016/j.intell.2008.04.001
Recently-derived variants of brain-size genes ASPM, MCPH1, CDK5RAP and BRCA1 not associated with general cognition, reading or language
Timothy C. Bates et al.
Abstract
Derived changes in genes associated with primary microcephaly (MCPH) have been suggested to be “currently sweeping to fixation” i.e., increasing in frequency in most populations, with the likely outcome that the derived allele will completely displace the ancestral allele over time. Possible causes for this sweep include effects on human reasoning and language. Here we test the hypothesis that these derived alleles are associated with current variation in spoken or written language and related traits. The association of derived alleles of the ASPM, MCPH1, CDK5RAP2 and BRCA1 genes was tested against well-validated measures of dyslexia, specific language impairment, working memory, IQ, and head-size in a family-based association study of over 1776 subjects from 789 families of twins. No evidence for association was found for any gene to any trait. The results strongly did not support the hypothesis that derived alleles in MCPH-related genes are related to the evolution of human language or cognition. Results were compatible with the alternate hypothesis, suggesting that adaptations in these genes associated with a dramatic increase in brain size have long since reached fixation and are now maintained by stabilizing selection.
Link
July 09, 2007
ASPM and the alphabet
Peter Frost who had proposed in the past a model for the evolution of light pigmentation in northern Europeans has come up with a new hypothesis linking the spread of the notorious ASPM gene variant with the spread of alphabetical writing.
Med Hypotheses. 2007 Jun 27; [Epub ahead of print]
The spread of alphabetical writing may have favored the latest variant of the ASPM gene.
Frost P.
Anthropology Department, C/o Bernard Saladin d’Anglure, Université Laval, Québec, Canada G1K 7P4.
ASPM, a gene that regulates brain growth, has evolved considerably in the primate lineage that leads to humans. It continued to evolve even after the emergence of modern humans, with the latest ASPM variant arising about 6000 years ago somewhere in the Middle East. The new variant then proliferated within and outside this region, reaching higher incidences in the Middle East (37-52%) and in Europe (38-50%) than in East Asia (0-25%). Despite its apparent selective advantage, this variant does not seem to improve cognitive performance, at least not on standard IQ tests. At present, we can only say that it probably assists performance on a task that exhibited the same geographic expansion from a Middle Eastern origin roughly 6000 years ago. The closest match seems to be the invention of alphabetical writing, specifically the task of transcribing speech and copying texts into alphabetical script. Though more easily learned than ideographs, alphabetical characters place higher demands on mental processing, especially under premodern conditions (continuous text with little or no punctuation, real-time stenography, absence of automated assistance for publishing or copying, etc.). This task was largely delegated to scribes of various sorts who enjoyed privileged status and probably superior reproductive success. Such individuals may have served as vectors for spreading the new ASPM variant.
Link
Med Hypotheses. 2007 Jun 27; [Epub ahead of print]
The spread of alphabetical writing may have favored the latest variant of the ASPM gene.
Frost P.
Anthropology Department, C/o Bernard Saladin d’Anglure, Université Laval, Québec, Canada G1K 7P4.
ASPM, a gene that regulates brain growth, has evolved considerably in the primate lineage that leads to humans. It continued to evolve even after the emergence of modern humans, with the latest ASPM variant arising about 6000 years ago somewhere in the Middle East. The new variant then proliferated within and outside this region, reaching higher incidences in the Middle East (37-52%) and in Europe (38-50%) than in East Asia (0-25%). Despite its apparent selective advantage, this variant does not seem to improve cognitive performance, at least not on standard IQ tests. At present, we can only say that it probably assists performance on a task that exhibited the same geographic expansion from a Middle Eastern origin roughly 6000 years ago. The closest match seems to be the invention of alphabetical writing, specifically the task of transcribing speech and copying texts into alphabetical script. Though more easily learned than ideographs, alphabetical characters place higher demands on mental processing, especially under premodern conditions (continuous text with little or no punctuation, real-time stenography, absence of automated assistance for publishing or copying, etc.). This task was largely delegated to scribes of various sorts who enjoyed privileged status and probably superior reproductive success. Such individuals may have served as vectors for spreading the new ASPM variant.
Link
April 20, 2007
Has ASPM been the target of recent selection?
Science Vol. 316. no. 5823, p. 370
Comment on "Ongoing Adaptive Evolution of ASPM, a Brain Size Determinant in Homo sapiens"
Fuli Yu et al.
Mekel-Bobrov et al. (Reports, 9 September 2005, p. 1720) suggested that ASPM, a gene associated with microcephaly, underwent natural selection within the last 500 to 14,100 years. Their analyses based on comparison with computer simulations indicated that ASPM had an unusual pattern of variation. However, when we compare ASPM empirically to a large number of other loci, its variation is not unusual and does not support selection.
Link
Comment on "Ongoing Adaptive Evolution of ASPM, a Brain Size Determinant in Homo sapiens"
Fuli Yu et al.
Mekel-Bobrov et al. (Reports, 9 September 2005, p. 1720) suggested that ASPM, a gene associated with microcephaly, underwent natural selection within the last 500 to 14,100 years. Their analyses based on comparison with computer simulations indicated that ASPM had an unusual pattern of variation. However, when we compare ASPM empirically to a large number of other loci, its variation is not unusual and does not support selection.
Link
January 16, 2007
ASPM and Microcephalin don't make people smarter
In Sep-05 a couple of papers showing that two brain genes, ASPM and Microcephalin had undergone positive selection in humans appeared in Science.
In May-06, another paper appeared that showed that Microcephalin and ASPM do not account for variability in brain size. This was a reasonable hypothesis to test, since these genes are implicated in microcephaly, a condition characterized by extremely small brains; it was thus natural to think that variation of these genes in normal subjects could also be correlated with their brain sizes; the data showed otherwise.
Now, a new study has appeared which studied the correlation of variants of these genes with IQ. As is well known, brain size only weakly correlates with IQ: in other words, by measuring how big a person's head is you get a very noisy estimate of how smart they are; other factors play a cumulatively bigger role. Since these genes are expressed in the brain, it might be possible that they worked their selective "magic" by conferring some advantage unrelated to brain size but related to IQ. However, even this is idea is also rejected by the data.
It remains to be seen what specific advantage -if any- the selected variants confer.
Hum Mol Genet. 2007 Jan 12; [Epub ahead of print]
The ongoing adaptive evolution of ASPM and Microcephalin is not explained by increased intelligence.
Mekel-Bobrov N et al.
Recent studies have made great strides towards identifying putative genetic events underlying the evolution of the human brain and its emergent cognitive capacities. One of the most intriguing findings is the recurrent identification of adaptive evolution in genes associated with primary microcephaly, a developmental disorder characterized by severe reduction in brain size and intelligence, reminiscent of the early hominid condition. This has led to the hypothesis that the adaptive evolution of these genes has contributed to the emergence of modern human cognition. As with other candidate loci, however, this hypothesis remains speculative due to the current lack of methodologies for characterizing the evolutionary function of these genes in humans. Two primary microcephaly genes, ASPM and Microcephalin, have been implicated not only in the adaptive evolution of the lineage leading to humans, but in ongoing selective sweeps in modern humans as well. The presence of both the putatively adaptive and neutral alleles at these loci provides a unique opportunity for using normal trait variation within humans to test the hypothesis that the recent selective sweeps are driven by an advantage in cognitive abilities. Here, we report a large-scale association study between the adaptive alleles of these genes and normal variation in several measures of IQ. Five independent samples were used, totaling 2,393 subjects, including both family-based and population-based datasets. Our overall findings do not support a detectable association between the recent adaptive evolution of either ASPM or Microcephalin and changes in IQ. As we enter the post-genomic era, with the number of candidate loci underlying human evolution growing rapidly, our findings highlight the importance of direct experimental validation in elucidating their evolutionary role in shaping the human phenotype.
Link
In May-06, another paper appeared that showed that Microcephalin and ASPM do not account for variability in brain size. This was a reasonable hypothesis to test, since these genes are implicated in microcephaly, a condition characterized by extremely small brains; it was thus natural to think that variation of these genes in normal subjects could also be correlated with their brain sizes; the data showed otherwise.
Now, a new study has appeared which studied the correlation of variants of these genes with IQ. As is well known, brain size only weakly correlates with IQ: in other words, by measuring how big a person's head is you get a very noisy estimate of how smart they are; other factors play a cumulatively bigger role. Since these genes are expressed in the brain, it might be possible that they worked their selective "magic" by conferring some advantage unrelated to brain size but related to IQ. However, even this is idea is also rejected by the data.
It remains to be seen what specific advantage -if any- the selected variants confer.
Hum Mol Genet. 2007 Jan 12; [Epub ahead of print]
The ongoing adaptive evolution of ASPM and Microcephalin is not explained by increased intelligence.
Mekel-Bobrov N et al.
Recent studies have made great strides towards identifying putative genetic events underlying the evolution of the human brain and its emergent cognitive capacities. One of the most intriguing findings is the recurrent identification of adaptive evolution in genes associated with primary microcephaly, a developmental disorder characterized by severe reduction in brain size and intelligence, reminiscent of the early hominid condition. This has led to the hypothesis that the adaptive evolution of these genes has contributed to the emergence of modern human cognition. As with other candidate loci, however, this hypothesis remains speculative due to the current lack of methodologies for characterizing the evolutionary function of these genes in humans. Two primary microcephaly genes, ASPM and Microcephalin, have been implicated not only in the adaptive evolution of the lineage leading to humans, but in ongoing selective sweeps in modern humans as well. The presence of both the putatively adaptive and neutral alleles at these loci provides a unique opportunity for using normal trait variation within humans to test the hypothesis that the recent selective sweeps are driven by an advantage in cognitive abilities. Here, we report a large-scale association study between the adaptive alleles of these genes and normal variation in several measures of IQ. Five independent samples were used, totaling 2,393 subjects, including both family-based and population-based datasets. Our overall findings do not support a detectable association between the recent adaptive evolution of either ASPM or Microcephalin and changes in IQ. As we enter the post-genomic era, with the number of candidate loci underlying human evolution growing rapidly, our findings highlight the importance of direct experimental validation in elucidating their evolutionary role in shaping the human phenotype.
Link
May 17, 2006
Microcephalin and ASPM do not Account for Brain Size Variability
Related to these recent papers, a new study on Microcephalin and ASPM, the "brain genes" under recent selection.
Human Molecular Genetics (Advance Access published online on May 10, 2006)
Normal Variants of Microcephalin and ASPM Do Not Account for Brain Size Variability
Roger P. Woods et al.
Abstract
Normal human brain volume is heritable. The genes responsible for variation in brain volume are not known. Microcephalin (MCPH1) and ASPM (abnormal spindle-like microcephaly associated) have been proposed as candidate genes since mutations in both genes are associated with microcephaly and common variants of each gene are apparently under strong positive selective pressure. In 120 normal subjects, we genotyped these variants and measured brain volumes using magnetic resonance imaging. We found no evidence that the selected alleles were associated with increases or decreases in brain volume. This result suggests that the selective pressure on these genes may be related to subtle neurobiological effects or to their expression outside the brain.
Link
Human Molecular Genetics (Advance Access published online on May 10, 2006)
Normal Variants of Microcephalin and ASPM Do Not Account for Brain Size Variability
Roger P. Woods et al.
Abstract
Normal human brain volume is heritable. The genes responsible for variation in brain volume are not known. Microcephalin (MCPH1) and ASPM (abnormal spindle-like microcephaly associated) have been proposed as candidate genes since mutations in both genes are associated with microcephaly and common variants of each gene are apparently under strong positive selective pressure. In 120 normal subjects, we genotyped these variants and measured brain volumes using magnetic resonance imaging. We found no evidence that the selected alleles were associated with increases or decreases in brain volume. This result suggests that the selective pressure on these genes may be related to subtle neurobiological effects or to their expression outside the brain.
Link
March 07, 2006
Recent positive selection in humans
Yet another study on positive selection in modern humans; you can find many many more by searching for "recent selection" or "positive selection" via the site search on the right sidebar.
The findings are reported in PLoS Biology (free text). Nicholas Wade also covers the study in the New York Times:
PLoS Biology Volume 4 | Issue 3 | MARCH 2006
A Map of Recent Positive Selection in the Human Genome
Benjamin F. Voight1, Sridhar Kudaravalli1, Xiaoquan Wen1, Jonathan K. Pritchard1*
The identification of signals of very recent positive selection provides information about the adaptation of modern humans to local conditions. We report here on a genome-wide scan for signals of very recent positive selection in favor of variants that have not yet reached fixation. We describe a new analytical method for scanning single nucleotide polymorphism (SNP) data for signals of recent selection, and apply this to data from the International HapMap Project. In all three continental groups we find widespread signals of recent positive selection. Most signals are region-specific, though a significant excess are shared across groups. Contrary to some earlier low resolution studies that suggested a paucity of recent selection in sub-Saharan Africans, we find that by some measures our strongest signals of selection are from the Yoruba population. Finally, since these signals indicate the existence of genetic variants that have substantially different fitnesses, they must indicate loci that are the source of significant phenotypic variation. Though the relevant phenotypes are generally not known, such loci should be of particular interest in mapping studies of complex traits. For this purpose we have developed a set of SNPs that can be used to tag the strongest ∼250 signals of recent selection in each population.
Link
The findings are reported in PLoS Biology (free text). Nicholas Wade also covers the study in the New York Times:
Providing the strongest evidence yet that humans are still evolving, researchers have detected some 700 regions of the human genome where genes appear to have been reshaped by natural selection, a principal force of evolution, within the last 5,000 to 15,000 years.From the new paper:
The genes that show this evolutionary change include some responsible for the senses of taste and smell, digestion, bone structure, skin color and brain function.
Many of these instances of selection may reflect the pressures that came to bear as people abandoned their hunting and gathering way of life for settlement and agriculture, a transition well under way in Europe and East Asia some 5,000 years ago.
A fully rigorous estimation of the ages of the candidate sweeps is difficult with the current data. However, making the simplistic assumption of a star-shaped genealogy for the favored haplotypes and assuming a generation time of 25 y, suggests average ages of ≍6,600 years and ≍10,800 years in the non-African, and African populations, respectively (Materials and Methods).
...
Some of the strongest signals of recent selection appear in various types of genes related to morphology. For example, four genes involved in skin pigmentation show clear evidence of selection in Europeans (OCA2, MYO5A, DTNBP1, TYRP1). All four genes are associated with Mendelian disorders that cause lighter pigmentation or albinism, and all are in different genomic locations, indicating the action of separate selective events. One of these genes, OCA2, is associated with the third longest haplotype on a high frequency SNP anywhere in the genome for Europeans. A fifth gene, SLC24A5, has recently been shown by another group to impact skin pigmentation and to have a derived, selected allele near fixation in Europeans [45]. Though iHS has reduced power for alleles near fixation, SNPs near this gene also show strong iHS signals in Europeans (Table S2).
Various genes involved in skeletal development have also been targets of recent selection. Three related proteins involved in bone morphogenesis show signals of selection in Europeans (BMP3 and BMPR2) and in East Asians (BMP5). In addition, GDF5, a gene in which mutations cause skeletal malformations, shows strong signals of selection in both Europeans and East Asians. Other morphological features also appear to be targets of selection, including hair formation and patterning in Yoruba (the keratin cluster near 17q12; and FZD6).
An important type of selective pressure that has confronted modern humans is the transition to novel food sources with the advent of agriculture and the colonization of new habitats [19,21]. As noted above, we see a strong signal of selection in the alcohol dehydrogenase (ADH) cluster in East Asians, including the third longest haplotype around a high frequency allele in East Asians. A variety of genes involved in carbohydrate metabolism have evidence for recent selection, including genes involved in metabolizing mannose (MAN2A1 in Yoruba and East Asians), sucrose (SI in East Asians), and lactose (LCT in Europeans). Processing of dietary fatty acids is another system with signals of strong selection, including uptake (SLC27A4 and PPARD in Europeans), oxidation (SLC25A20 in East Asians) and regulation (NCOA1 in Yoruba and LEPR in East Asians). The latter gene (LEPR) is the leptin receptor and plays an important role in regulating adipose tissue mass.
Recent articles have proposed that genes involved in brain development and function may have been important targets of selection in recent human evolution [8,9]. While we do not find evidence for selection in the two genes reported in those studies (MCPH1 and ASPM), we do find signals in two other microcephaly genes, namely, CDK5RAP2 in Yoruba, and CENPJ in Europeans and East Asians [46]. Though there is not an overall enrichment for neurological genes in our gene ontology analysis, several other important brain genes also have signals of selection, including the primary inhibitory neurotransmitter GABRA4, an Alzheimer's susceptibility gene PSEN1, and SYT1 in Yoruba; the serotonin transporter SLC6A4 in Europeans and East Asians; and the dystrophin binding gene SNTG1 in all populations.
PLoS Biology Volume 4 | Issue 3 | MARCH 2006
A Map of Recent Positive Selection in the Human Genome
Benjamin F. Voight1, Sridhar Kudaravalli1, Xiaoquan Wen1, Jonathan K. Pritchard1*
The identification of signals of very recent positive selection provides information about the adaptation of modern humans to local conditions. We report here on a genome-wide scan for signals of very recent positive selection in favor of variants that have not yet reached fixation. We describe a new analytical method for scanning single nucleotide polymorphism (SNP) data for signals of recent selection, and apply this to data from the International HapMap Project. In all three continental groups we find widespread signals of recent positive selection. Most signals are region-specific, though a significant excess are shared across groups. Contrary to some earlier low resolution studies that suggested a paucity of recent selection in sub-Saharan Africans, we find that by some measures our strongest signals of selection are from the Yoruba population. Finally, since these signals indicate the existence of genetic variants that have substantially different fitnesses, they must indicate loci that are the source of significant phenotypic variation. Though the relevant phenotypes are generally not known, such loci should be of particular interest in mapping studies of complex traits. For this purpose we have developed a set of SNPs that can be used to tag the strongest ∼250 signals of recent selection in each population.
Link
February 03, 2006
More positive selection in recent humans: CASP12
It seems that positive selection studies are coming out in droves; you can use the search function ("positive selection") to find a bunch of them reported on this blog.
The newest study appears as a preprint in the American Journal of Human Genetics and is about a gene called human caspase-12 (CASP12) which has two variants: the active one is the original form, but the inactive one seems to have undergone positive selection beginning 60-100ky ago.
It is suggested that the inactive form confers some advantage against sepsis, which was also implicated in a frequent mtDNA haplogroup recently.
The distribution of the two versions of the gene...
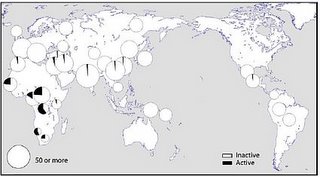
... shows that the active version is largely confined to Sub-Saharan Africa. This parallels the situation in Microcephalin and ASPM.
The highest frequencies of the active version are in the Mbuti (0.60) and the San (0.57). These populations have a high frequency of non-M168 related Y-chromosomes and non-L3 related mtDNA and are thus descended from the most ancient African populations (which I have called "Paleoafricans"), rather than the more recent emergence of an population in east Africa ("Afrasians") which spawned the Eurasians and assimilated most Paleoafricans during its spread within Africa. In this regard, it is interesting that an east African population from Kenya shows the minimum (0.04) frequency of the active gene of CASP12.
American Journal of Human Genetics (in press)
Spread of an inactive form of caspase-12 in humans due to recent
positive selection
Yali Xue et al.
Abstract
The human caspase-12 gene is polymorphic for the presence or absence of a stop codon, resulting in the occurrence of both active (ancestral) and inactive (derived) forms of the gene in the population. It has previously been shown that carriers of the inactive gene are more resistant to severe sepsis. We have now investigated whether the inactive form has spread because of neutral drift or positive selection. We determined its distribution in a worldwide sample of 52 populations and re-sequenced the gene in 77 individuals from the HapMap Yoruba, Han Chinese and European populations. There is strong evidence for positive selection from low diversity, skewed allele frequency spectra and the predominance of a single haplotype. We suggest that the inactive form of the gene arose in Africa ~100-500 thousand years ago (KYA) and was initially neutral or almost neutral, but that positive selection beginning ~60-100 KYA drove it to near-fixation. We further propose that its selective advantage was sepsis resistance in populations that experienced more infectious diseases as population sizes and densities increased.
Link (pdf)
September 09, 2005
Stop the presses... huge papers on brain evolution in recent humans
I am very sure that a set of new papers (one two) in Science will generate a huge amount of buzz. I will blog in more detail about them later, but for now, this Red Nova story covers the findings pretty well. The interesting part:

Fig. 3. Global frequencies of Microcephalin haplogroup D chromosomes (defined as having the derived C allele at the G37995C diagnostic SNP) in a panel of 1184 individuals.

Fig. 1. Worldwide frequencies of ASPM haplogroup D chromosomes (defined as having the derived G allele at the A44871G diagnostic polymorphism), based on a panel of 1186 individuals.
UPDATE
Here is what these studies mean:
In a recent article, Erik Trinkaus has surveyed the human paleoanthropological record, and wrote that:
The question is: why did the new variant not get selected in Sub-Saharan Africans? There are only two possible explanations:
Now, it is well known that the greatest difference in modern human genetic variation is between Sub-Saharan Africans and non-Sub-Saharan Africans. This is the result of the fact that humans originated in Africa, and possess only a subset of the variation that exists there.
As I have written before, there is good reason to believe that a human group originated in eastern Africa ("Afrasians") and came to colonize the rest of the world in relatively recent times. But, the rest of the African continent was already inhabited by pre-existing anatomically modern humans ("Paleoafricans") since at least 150,000 years in the past. These "Paleoafricans" were separated from the "Afrasians", as evidenced by the fact that typical "Paleoafrican" markers, originating long before the 40,000BP cutoff date, such as Y-haplogroups A and B and mtDNA haplogroups L0-L2 are not found in Eurasia.
If my theory is correct, then we don't need to propose some unquantifiable peculiarity of the African environment. Rather, the new Microcephalin variant has a low frequency in Sub-Saharan Africans precisely because it emerged in the Afrasians of eastern Africa that started colonizing the world around 40,000BP and was later added to the Paleoafrican populations of Sub-Saharan Africa. It simply has not had enough time to spread in most of Africa!
The second variant (of ASPM) is even more impressive, because it started to spread only 5,800 years ago, although the confidence margins are wide. The only movement which could have affected so many populations of Eurasia, regardless of language, in the last few millennia is the Neolithic expansion, followed by population growth in the first civilizations of the Near East and China.
It seems all by certain that the variant first appeared in Western Eurasia. It could have been carried easily to the east by the Near Eastern Neolithic people who reached India. It would only take a small step to make the jump to the Mongoloid world; once introduced into the population, it would also undergo the same selection process that made it so frequent among Caucasoids. However, agriculture begins much later among Mongoloids and even later among Australoids. So, the low frequency of the new variant in these populations is a consequence of the fact that it has had less time to spread among these populations.
The new ASPM variant is lacking in Sub-Saharan Africans and Native Americans. These results can be easily explained:
Science, Vol. 309 No. 5741
Microcephalin, a Gene Regulating Brain Size, Continues to Evolve Adaptively in Humans
Patrick D. Evans et al.
The gene Microcephalin (MCPH1) regulates brain size and has evolved under strong positive selection in the human evolutionary lineage. We show that one genetic variant of Microcephalin in modern humans, which arose ~37,000 years ago, increased in frequency too rapidly to be compatible with neutral drift. This indicates that it has spread under strong positive selection, although the exact nature of the selection is unknown. The finding that an important brain gene has continued to evolve adaptively in anatomically modern humans suggests the ongoing evolutionary plasticity of the human brain. It also makes Microcephalin an attractive candidate locus for studying the genetics of human variation in brain-related phenotypes.
Link
Ongoing Adaptive Evolution of ASPM, a Brain Size Determinant in Homo sapiens
Nitzan Mekel-Bobrov et al.
The gene ASPM (abnormal spindle-like microcephaly associated) is a specific regulator of brain size, and its evolution in the lineage leading to Homo sapiens was driven by strong positive selection. Here, we show that one genetic variant of ASPM in humans arose merely about 5800 years ago and has since swept to high frequency under strong positive selection. These findings, especially the remarkably young age of the positively selected variant, suggest that the human brain is still undergoing rapid adaptive evolution.
Link
The team also observed geographic differences. For haplogroup D of ASPM, they found that it occurs more frequently in Europeans and surrounding populations including, North Africans, Middle Easterners, and South Asians, and at a lower incidence in East Asians, New World Indians and sub-Saharan Africans. For microcephalin, the researchers found that haplogroup D is more abundant in populations outside of sub-Saharan Africa.For now, from the papers:

Fig. 3. Global frequencies of Microcephalin haplogroup D chromosomes (defined as having the derived C allele at the G37995C diagnostic SNP) in a panel of 1184 individuals.

Fig. 1. Worldwide frequencies of ASPM haplogroup D chromosomes (defined as having the derived G allele at the A44871G diagnostic polymorphism), based on a panel of 1186 individuals.
UPDATE
Here is what these studies mean:
- Microcephalin and ASPM are genes involved in regulating brain size
- A variant of Microcephalin has reached very high frequencies in non-Sub-Saharan Africans in the last 37,000 years.
- A variant of ASPM has reached very high frequencies especially in Caucasoids but also in some southern Mongoloids and Australoids in the last 5,800 years.
- It is almost inconceivable that these two factors were caused by random factors (drift). Therefore selection has acted on these two genes, favoring the new Microcephalin variant in non-Sub-Saharan Africans and the ASPM especially in Caucasoids, but also to a lesser extent in some southern Mongoloid and Australoid groups.
- We know absolutely nothing about what the new Microcephalin and ASPM variants actually do. What we do know is that they confer some substantial advantage that has caused them to grow in numbers. Perhaps, they confer some cognitive or behavioral ability.
In a recent article, Erik Trinkaus has surveyed the human paleoanthropological record, and wrote that:
The earliest candidates for human anatomical modernity, those between ca. 150,000 and 195,000 years B.P. in Africa, are best considered as bridging a morphological gap between late archaic and early modern humans.Now, it may be a coincidence that the spread of humans throughout Africa and into Eurasia happened at around the same time that the new Microcephalin variant appeared, but the timing is certainly suggestive.
...
The spread of modern humans thoughout Africa and into Eurasia occurred after 50,000 years B.P. and probably after 40,000 years B.P., 100,000 years after their appearance.
The question is: why did the new variant not get selected in Sub-Saharan Africans? There are only two possible explanations:
- There is something in the Sub-Saharan African environment which did not allow the variant to be selected; in other words: the variant did not confer an advantage in Africa itself.
- The gene pool of most Sub-Saharan Africans did not possess the new Microcephalin variant. Hence the variant did not get selected because it was lacking in the Sub-Saharan African gene pool.
Now, it is well known that the greatest difference in modern human genetic variation is between Sub-Saharan Africans and non-Sub-Saharan Africans. This is the result of the fact that humans originated in Africa, and possess only a subset of the variation that exists there.
As I have written before, there is good reason to believe that a human group originated in eastern Africa ("Afrasians") and came to colonize the rest of the world in relatively recent times. But, the rest of the African continent was already inhabited by pre-existing anatomically modern humans ("Paleoafricans") since at least 150,000 years in the past. These "Paleoafricans" were separated from the "Afrasians", as evidenced by the fact that typical "Paleoafrican" markers, originating long before the 40,000BP cutoff date, such as Y-haplogroups A and B and mtDNA haplogroups L0-L2 are not found in Eurasia.
If my theory is correct, then we don't need to propose some unquantifiable peculiarity of the African environment. Rather, the new Microcephalin variant has a low frequency in Sub-Saharan Africans precisely because it emerged in the Afrasians of eastern Africa that started colonizing the world around 40,000BP and was later added to the Paleoafrican populations of Sub-Saharan Africa. It simply has not had enough time to spread in most of Africa!
The second variant (of ASPM) is even more impressive, because it started to spread only 5,800 years ago, although the confidence margins are wide. The only movement which could have affected so many populations of Eurasia, regardless of language, in the last few millennia is the Neolithic expansion, followed by population growth in the first civilizations of the Near East and China.
It seems all by certain that the variant first appeared in Western Eurasia. It could have been carried easily to the east by the Near Eastern Neolithic people who reached India. It would only take a small step to make the jump to the Mongoloid world; once introduced into the population, it would also undergo the same selection process that made it so frequent among Caucasoids. However, agriculture begins much later among Mongoloids and even later among Australoids. So, the low frequency of the new variant in these populations is a consequence of the fact that it has had less time to spread among these populations.
The new ASPM variant is lacking in Sub-Saharan Africans and Native Americans. These results can be easily explained:
- Sub-Saharan African agriculture is late, and moreover there has been almost no gene flow from Eurasia into Sub-Saharan Africa, with a few occasional exceptions. So, the ASPM variant did not exist in the Sub-Saharan African gene pool, and could thus have not been selected.
- Native Americans migrated into the New World in Paleolithic times. Naturally, the ASPM variant was not present in their ancestral gene pool yet, so it could not have been selected.
Science, Vol. 309 No. 5741
Microcephalin, a Gene Regulating Brain Size, Continues to Evolve Adaptively in Humans
Patrick D. Evans et al.
The gene Microcephalin (MCPH1) regulates brain size and has evolved under strong positive selection in the human evolutionary lineage. We show that one genetic variant of Microcephalin in modern humans, which arose ~37,000 years ago, increased in frequency too rapidly to be compatible with neutral drift. This indicates that it has spread under strong positive selection, although the exact nature of the selection is unknown. The finding that an important brain gene has continued to evolve adaptively in anatomically modern humans suggests the ongoing evolutionary plasticity of the human brain. It also makes Microcephalin an attractive candidate locus for studying the genetics of human variation in brain-related phenotypes.
Link
Ongoing Adaptive Evolution of ASPM, a Brain Size Determinant in Homo sapiens
Nitzan Mekel-Bobrov et al.
The gene ASPM (abnormal spindle-like microcephaly associated) is a specific regulator of brain size, and its evolution in the lineage leading to Homo sapiens was driven by strong positive selection. Here, we show that one genetic variant of ASPM in humans arose merely about 5800 years ago and has since swept to high frequency under strong positive selection. These findings, especially the remarkably young age of the positively selected variant, suggest that the human brain is still undergoing rapid adaptive evolution.
Link
Subscribe to:
Posts (Atom)
