 From the paper:
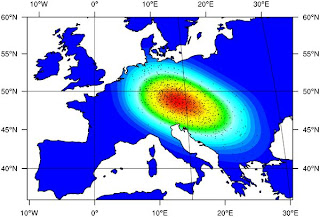
From the paper:Following acceptance at the 0.5% level and regression adjustment we found that the most probable location where an LP allele first underwent selection among dairying farmers lies in a region between the central Balkans and central Europe (see Figure 3). It should be noted that, as simulated, we did not attempt to identify the location where the LP −13,910*T allele first arose. Instead we assumed that it started to rise to appreciable frequencies only after selection began among dairying farmers, initially at the particular location we estimated. The timing of the start of this gene-culture coevolution process was therefore strongly influenced by the arrival time of dairying farmers at the location where selection began in simulations. Since we selected simulations that give a good fit to the timing of the arrival of farming at different locations [31], we estimated a narrow range of dates for when selection began (95% CI 6,256 to 8,683 years BP;In other words, selection for the lactase persistence allele did not result in modern Europeans having a larger proportion of their ancestry from the place where this process began.
...
Although not strictly a parameter of the model presented we have applied the ABC approach to estimate the genetic contribution of people living in the deme where LP-dairying gene-culture coevolution began, and its 8 surrounding demes, to the modern European gene-pool (95% CI 2.83 to 27.4%; mode = 7.47%; see Figure 4B) ... We then compared the distributions of genetic contribution (of people living in and around the LP-dairying start deme to the modern European genepool) with and without selection acting. To our surprise the two distributions are nearly identical.
From the paper:
Perhaps the most interesting result presented here is our estimation of the geographic and temporal origins of LP-dairying co-evolution. We find the highest posterior probabilities for a region between the central Balkans and central Europe (see Figure 3). At first sight such a location of origin may seem counter intuitive since it is far-removed from Northwest Europe, where the −13,910*T allele is found at highest frequency. However, previous simulations have shown that the geographic centroid of allele can be offset from its location of origin, particularly when it occurs on the wave front of a demographic expansion [29],[30]. The lactase-dairying coevolution origin region inferred here is consistent with a number of archaeologically attested patterns concerning the emergence and spread of dairying. Recent carbon isotope ratios from lipids extracted from archaeological sherds show the presence of milk fats in present-day western Turkey and connect these findings to an increased importance of cattle herding [26], [45]–[48]. In general, the spread of the Neolithic lifestyle from the Aegean to Central Europe goes hand in hand with the decline of the importance of sheep and goat and the rise in frequency of cattle bones in archaeological assemblages. While the Balkans at the beginning of the Neolithic still shows a variety of subsistence strategies [49], the middle Neolithic in SE-Europe and the earliest Neolithic in Central Europe after 7,500 BP show a clear preponderance of cattle.
A related recent study: Demography and selection in the spread of Lactase persistence in Europe (Gerbault et al. 2009)
UPDATE (Aug 29):
John Hawks raises two objections to the current paper:
There's only one little problem: It's hard to see how the same scenario gets the allele to India. Or, for that matter, Ireland. The authors posit that Indian lactase persistence will be found to be caused by a "diversity" of alleles. They seem to have missed this paper that found a greater diversity of lactase-associated haplotypes "north of the Caucasus" -- consistent with an initial steppe dispersal. OK, that's two problems, and they're not little.
I don't really see a problem with the spread of the allele to Ireland or to India. What the authors of this paper claim is that the allele began to be selected in Central Europe, not that it originated there. Its presence in Ireland or India does not strictly require any population movements from Central Europe. But there is also a plausible case for gene flow from Central Europe to either direction (Celts in the case of Ireland, and small-scale European admixture routinely detected in admixture studies that include South Asian populations).
As for the cited paper, it completely lacks samples from Central Europe, the Balkans, and Anatolia, hence its conclusion that the allele originated "north of the Caucasus" is spurious, and is not incompatible with the current paper which proposes a Balkan/Central European beginning of its selection process.
UPDATE (Aug 31)
John Hawks suggests in the comments that inclusion of South Asia into the model would shift the place of origin of the allele towards the east, and away from Central Europe. I do agree that a full model should account for the presence of allele as far as India or Central Asia. However, I doubt that their inclusion would have a major effect, for two reasons:
- Higher allele frequency in northwestern Europe compared to India suggests that the "point of origin" ought to be closer to the former than to the latter, or that the allele's selection began earlier in the former than in the latter.
- We must account for terrain and mode of transmission. The steppelands stretching from eastern Europe to the outskirts of China, combined with the invention of full pastoral nomadism made it possible for the spread of genes at a speed impossible for regular "demic diffusion". Moreover, a great part of this territory was essentially devoid of previous populations, and, the economy of the nomads necessitated its continued positive selection. Thus, the allele's frequency would not have been diluted by the time it reached the eastern ends of its expansion.
The opposite trip (introduction to Europe from eastern European nomads) is also possible, but there are reasons to doubt this:
- The beginning of selection inferred in the current study is much older than the invention of pastoral nomadism. Inclusion of more populations could only push the time further into the past; it could not make it more recent. Thus, advocates of an "eastern" solution must explain how an allele appears to have started experiencing selection in the geographical region examined in the current paper thousands of years before it was introduced from the east.
- An eastern-western mode of transmission would result in an eastern-western cline, not a northern-southern one. An additional mechanism would need to be invoked to explain the latter.
PLoS Comput Biol 5(8): e1000491. doi:10.1371/journal.pcbi.1000491
The Origins of Lactase Persistence in Europe
Yuval Itan et al.
Abstract
Lactase persistence (LP) is common among people of European ancestry, but with the exception of some African, Middle Eastern and southern Asian groups, is rare or absent elsewhere in the world. Lactase gene haplotype conservation around a polymorphism strongly associated with LP in Europeans (−13,910 C/T) indicates that the derived allele is recent in origin and has been subject to strong positive selection. Furthermore, ancient DNA work has shown that the −13,910*T (derived) allele was very rare or absent in early Neolithic central Europeans. It is unlikely that LP would provide a selective advantage without a supply of fresh milk, and this has lead to a gene-culture coevolutionary model where lactase persistence is only favoured in cultures practicing dairying, and dairying is more favoured in lactase persistent populations. We have developed a flexible demic computer simulation model to explore the spread of lactase persistence, dairying, other subsistence practices and unlinked genetic markers in Europe and western Asia's geographic space. Using data on −13,910*T allele frequency and farming arrival dates across Europe, and approximate Bayesian computation to estimate parameters of interest, we infer that the −13,910*T allele first underwent selection among dairying farmers around 7,500 years ago in a region between the central Balkans and central Europe, possibly in association with the dissemination of the Neolithic Linearbandkeramik culture over Central Europe. Furthermore, our results suggest that natural selection favouring a lactase persistence allele was not higher in northern latitudes through an increased requirement for dietary vitamin D. Our results provide a coherent and spatially explicit picture of the coevolution of lactase persistence and dairying in Europe.
Link

7 comments:
It seems to me that this is in line with what I said. Two observations: 1) going to North-West they were undoubtedly R1b1b2 and this contrasts with who thinks R1b1b2 younger; 2) they, following the given theories, didn't speak Indo-European if the split of the Hittite happened on 6,000YBP: then many people have said many foolishnesses.
The Authors write: “We infer that the coevolution of European LP and dairying
originated in a region between central Europe and the northern
Balkans around 6,256 to 8,683 years BP. We propose the following
scenario: after the arrival of the Neolithic in south-eastern Europe
and the increasing importance of cattle herding and dairying,
natural selection started to act on a few LP individuals of the early
Neolithic cultures of the northern Balkans. After the initial slow
increase of LP frequency in those populations and the onset of the
Central European LBK culture around 7,500 BP, LP frequencies
rose more rapidly in a gene-culture co-evolutionary process and on
the wave front of a demographic expansion, leading to the establishment of highly
developed cattle- (and partly also goat-) based dairying economies
during the Middle Neolithic of central Europe around 6,500 BP. A
latitudinal effect on selection for LP, through an increased
requirement for dietary vitamin D, is unnecessary to explain
the high frequencies found in northern Europe”.
I propose, like I have supported in the past, that those people was R1b1b2-L23+
coming from Italy, where this haplotype has the high frequency (30% of all R1b
in the Argiedude’s spreadsheet) and all the previous haplotypes. They were
hunter-gatherers converted to agriculture. This explains the high percentage of
R-L23+ in Central Europe (Czech Republic) but not elsewhere. Of course among them
there were some subclades: probably R-L51 and R-P312 and the others must be ascertained.
It is difficult in fact to explain all the Italian R-U152 by a back migration from Central Europe
with Celts.
I am a natural skeptic. The study says LP in Africans and Middle Easterners is due to a different mutation on a similar site to that found in Europeans.
The people according to theory who introduced farming to Europe, including animal husbandry, were Near Easterners i.e from the Middle East. These Middle Easterners had domesticated both the goat and sheep prior to bovines. All impregnated female mammalian animals produced milk, and all these milks contain Milk sugar, Lactose. It begs belief to accept that the Middle Eastern farmers with their sheep, goats, bovines and wheat did not use milk and started the selection process to be able to process and digest Lactose in milk without producing harsh physiological effects. The dates in question for the selection of the gene for LP is within the Neolithic age and can't be pinpointed to any particular Neolithic age Middle Eastern group of European economic refugees. As I said I am skeptical. Anyway if LP was so advantageous which I doubt*, it would be nearly total in all European groups as all European groups practice dairying. Dairying is very expensive and labour intensive. In my country most farmers are being forced out of dairying as it is not economically viable. In the Neolithic age, sheep and goats would have been better economically than bovines, and the production of grains better for the survival of farming communities with some minor hunting for meat and fish.
* You have to factor in zoonoses like Smallpox, Measles, Tuberculosis which bovines and their milk are the original source into human populations. Drinking unprocessed milk could kill you.
But there is also a plausible case for gene flow from Central Europe to either direction (Celts in the case of Ireland, and small-scale European admixture routinely detected in admixture studies that include South Asian populations).
Oh, I agree that the allele could have gotten there; it just couldn't have gotten there under the authors' model. The allele frequency cline in Europe is the only thing they try to fit to their model; putting a nonzero frequency in Pakistan under their model would require an origin much farther to the east.
It is certainly an omission, but not a very significant one in terms of the overall conclusion.
The steppe region has a mode of transmission peculiar to it, namely full pastoral nomadism. Once the allele gets to eastern Europe, the rest of the trip is -by comparison- free, and the presence of the selected allele in India/Pakistan or even Mongolia is easily explained.
The possible introduction of the allele from the east into Europe may need to be modeled separately, but then you'd have to make the whole evolution of lactase persistence take place in much less time (clearly incompatible with the dates inferred here), and to turn an east-west cline into a predominantly northwestern-southeastern one.
The author was on BBC radio he says those able to digest milk had a big advantage but for reasons unconnected with vitamin D {which would rule out calcium}. So lactase persistence surfed a wave of expansion north but it had no special advantage there.
By the way milk does not naturally contain a nutritionally significant level of vitamin D. Even fish has far less than most people think
An Evaluation of the Vitamin D3 Content in Fish: Is the Vitamin D Content Adequate to Satisfy the Dietary Requirement for Vitamin D?
"It has been assumed that fish, especially oily fish such as salmon, mackerel and blue fish are excellent sources of vitamin D3. However, our analysis of the vitamin D content in a variety of fish species that were thought to contain an adequate amount of vitamin D did not have an amount of vitamin D that is listed in food charts. There needs to be a reevaluation of the vitamin D content in foods that have been traditionally recommended as good sources of naturally occurring vitamin D."
Eh, that paper adds little new.
Fact is that it is difficult to get sufficient vitamin D in most diets, without sufficient exposure to sun, and sufficiently light skin color. Inuits traditionally could afford to have dark skin color, because so much of their diet is seafood and sea mammals (that feed of seafood).
Hunting does not really help; early central/northern Europeans (while having fish as only part of their diet) likely already required to have relatively light skin color, except for some post-LGM coastal populations that had major seafood sustenance.
This is a better overview:
http://ods.od.nih.gov/factsheets/vitamind.asp
Post a Comment