June 30, 2010
Earliest copper smelting from the Balkans
On the Origins of Extractive Metallurgy: New evidence from Europe
Miljana Radivojević et al.
The beginnings of extractive metallurgy in Eurasia are contentious. The first cast copper objects in this region emerge c7000 years ago, and their production has been tentatively linked to centres in the Near East. This assumption, however, is not substantiated by evidence for copper smelting in those centres. Here, we present results from recent excavations from Belovode, a Vinča culture site in Eastern Serbia, which has provided the earliest direct evidence for copper smelting to date. The earliest copper smelting activities there took place c7000 years ago, contemporary with the emergence of the first cast copper objects. Through optical, chemical and provenance analyses of copper slag, minerals, ores and artefacts, we demonstrate the presence of an established metallurgical technology during this period, exploiting multiple sources for raw materials. These results extend the known record of copper smelting by more than half a millennium, with substantial implications. Extractive metallurgy occurs at a location far away from the Near East, challenging the traditional model of a single origin of metallurgy and reviving the possibility of multiple, independent inventions.
Link
June 28, 2010
Half of hidden heritability found (for height, at least)
This bears great promise as it may hint that genome-wide association studies, that have come under substantial criticism lately, may be failing not because of an inherent flaw, but rather because they are not sampling enough individuals.
The discovered SNPs account for 45% of the heritability of height. Where is the rest? The authors argue for two additional sources:
First, SNPs in current microarray chips sample the genome incompletely. Locations in-between discovered SNPs are in incomplete linkage disequilibrium with the discovered SNPs. So, there is undetected polymorphism, in the gaps between the hundreds of thousands of SNPs in current chips, that may explain a portion of the missing heritability.
Second, SNPs have different minor allele frequencies. For example, in one SNP the minor allele may occur at 10% of individuals, while in others at 30%. This is important, because it is more difficult to arrive at a statistically significant result in the former case.
Consider a SNP with a minor allele frequency of 2%. Then, if you sample 1,000 individuals, only about 20 of them are expected to have the minor allele. You cannot estimate the average height of the minor allele with a sample of 20 people as securely as you can with a sample of 500. Thus, if the SNP influences height in a small way, you will not be able to detect it.
A further complication, which I've written about before, is that some variation in the human genome is family-related, or at least occurs at fewer individuals than the allele frequency cutoff. If 99.9% of people have C at a given location and 0.1% of people have T, this variant is unlikely to be included in a microarray chp, because it is too rare to matter economically: you would only get a handful of individuals -if you're lucky- in a sample of 1,000 for such a variant. However, rarity does not mean that the variant is functionally unimportant, and the rare allele may play a substantial role in the height of the people who possess it.
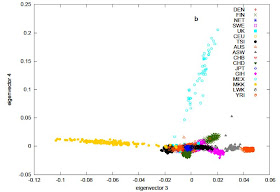
In the European-only PCA we see the familiar north-south gradient (anchored by Tuscans TSI and Netherlands NET on either side), and the orthogonal deviation of the Finns. Swedes (SWE) occupy a northern European end of the spectrum like the Dutch, but are spread towards Finns, reflecting low-level Finnish admixture in that population. Conversely, Finns are variable along the same axis, reflecting variable levels of admixture. Australians (AUS) and UK, on the other hand, are on the northern European edge of the main European gradient, with a number of individuals spread toward the Tuscan side.
 The PCA with all populations is also quite interesting. East Eurasians (Chinese and Japanese) form a tight pole at the bottom right. Gujarati Indians (GIH) form a different pole, spread towards Europeans, reflecting variable levels of West Eurasian admixture in that population, probably corresponding to the ANI element recently discovered in Indian populations. Mexicans (MEX) are spread towards East Asians, reflecting their Amerindian admixture, but notice how they are not positioned exactly on the European-East Asian axis, probably reflecting the third, minority, Sub-Saharan element in their ancestry, as well as the fact that Amerindians are not perfectly represented by East Asians. Finns are tilted towards East Asians, as expected, reflecting the fact that their genetic specificity vis a vis Northern Europeans is due to low-level East Eurasian ancestry.
The PCA with all populations is also quite interesting. East Eurasians (Chinese and Japanese) form a tight pole at the bottom right. Gujarati Indians (GIH) form a different pole, spread towards Europeans, reflecting variable levels of West Eurasian admixture in that population, probably corresponding to the ANI element recently discovered in Indian populations. Mexicans (MEX) are spread towards East Asians, reflecting their Amerindian admixture, but notice how they are not positioned exactly on the European-East Asian axis, probably reflecting the third, minority, Sub-Saharan element in their ancestry, as well as the fact that Amerindians are not perfectly represented by East Asians. Finns are tilted towards East Asians, as expected, reflecting the fact that their genetic specificity vis a vis Northern Europeans is due to low-level East Eurasian ancestry. An interesting aspect of the first two PCs is the fact that the Maasai (MKK) and Luhya (LUW) from Kenya are not separated from Caucasoids, and neither are Yoruba from Nigeria (YRI). This is a good reminder of the fact that identity in the first two principal components may mask difference revealed in higher order components. This difference (at least for Maasai) is seen in the next two PCs.
An interesting aspect of the first two PCs is the fact that the Maasai (MKK) and Luhya (LUW) from Kenya are not separated from Caucasoids, and neither are Yoruba from Nigeria (YRI). This is a good reminder of the fact that identity in the first two principal components may mask difference revealed in higher order components. This difference (at least for Maasai) is seen in the next two PCs.Common SNPs explain a large proportion of the heritability for human height
Jian Yang et al.
Abstract
SNPs discovered by genome-wide association studies (GWASs) account for only a small fraction of the genetic variation of complex traits in human populations. Where is the remaining heritability? We estimated the proportion of variance for human height explained by 294,831 SNPs genotyped on 3,925 unrelated individuals using a linear model analysis, and validated the estimation method with simulations based on the observed genotype data. We show that 45% of variance can be explained by considering all SNPs simultaneously. Thus, most of the heritability is not missing but has not previously been detected because the individual effects are too small to pass stringent significance tests. We provide evidence that the remaining heritability is due to incomplete linkage disequilibrium between causal variants and genotyped SNPs, exacerbated by causal variants having lower minor allele frequency than the SNPs explored to date.
Link
Genetic structure of Qatar (Hunter-Zinck et al. 2010)
 (Last Update Jul 11)
(Last Update Jul 11)A Mantel test comparing Qatari subgroups and surname origins indicates highly significant (p = 0.0001) correlations across the three population groups in the frequency of these name classifications, with the Qatar1 population having mostly Arab surnames, the Qatar2 population having a large Persian component, and the Qatar3 population appearing to be the most diverse and having the largest African component (Figure 7).
doi:10.1016/j.ajhg.2010.05.018
Population Genetic Structure of the People of Qatar
Haley Hunter-Zinck et al.
People of the Qatar peninsula represent a relatively recent founding by a small number of families from three tribes of the Arabian Peninsula, Persia, and Oman, with indications of African admixture. To assess the roles of both this founding effect and the customary first-cousin marriages among the ancestral Islamic populations in Qatar's population genetic structure, we obtained and genotyped with Affymetrix 500k SNP arrays DNA samples from 168 self-reported Qatari nationals sampled from Doha, Qatar. Principal components analysis was performed along with samples from the Human Genetic Diversity Project data set, revealing three clear clusters of genotypes whose proximity to other human population samples is consistent with Arabian origin, a more eastern or Persian origin, and individuals with African admixture. The extent of linkage disequilibrium (LD) is greater than that of African populations, and runs of homozygosity in some individuals reflect substantial consanguinity. However, the variance in runs of homozygosity is exceptionally high, and the degree of identity-by-descent sharing generally appears to be lower than expected for a population in which nearly half of marriages are between first cousins. Despite the fact that the SNPs of the Affymetrix 500k chip were ascertained with a bias toward SNPs common in Europeans, the data strongly support the notion that the Qatari population could provide a valuable resource for the mapping of genes associated with complex disorders and that tests of pairwise interactions are particularly empowered by populations with elevated LD like the Qatari.
Link
June 25, 2010
Y chromosome haplogroup I and disease-based selection
In this study, we aimed to refine our understanding of AIDS susceptibility associated with hg-I. We first analyzed AIDS progression differences between hg-I subhaplogroups and the phylogenetically closest haplogroup, haplogroup J (hg-J). Previously, we reported faster progression to AIDS outcomes in hg-I compared to the most common European haplogroup R, but did not analyze hg-J. As hg-I and hg-J are sister clades with similar time depths, and likely to have relatively closely related sequence organizations, it is interesting to see if the disease susceptibility is unique to hg-I or common among the hg-IJ clades.
There were significant differences between the hg-I subhaplogroups and hg-J samples for AIDS progression, where hg-I subhaplogroups depleted CD4+ T cells and progressed to AIDS 1993 and AIDS 1987 faster (Table 1). However, the faster AIDS progression signal (compared to the hg-J samples) was not significant for every individual hg-I subhaplogroup, probably due to small sample sizes resulting in lack of statistical power. When all hg-I subhaplogroups were combined (representing hg-I) and compared against hg-J, the significant AIDS progression differences were more evident (Pp0.02 for each AIDS end point; Table 1).In contrast, when the hg-I subhaplogroup samples were compared against each other, the Cox analyses did not show a significant AIDS progression differences between them (Table 1).
- Y chromosome haplogroup I and rapid HIV progression
- mtDNA haplogroups and AIDS progression
- Male infertility induced by mtDNA/Y unfavorable combination? An association study on human mitochondrial DNA
Abstract
We attempted to refine the understanding of an association of Y-chromosomal haplogroup I (hg-I) with enhanced AIDS progression that had been previously reported. First, we compared the progression phenotype between hg-I and its phylogenetically closest haplogroup J. Then, we took a candidate gene approach resequencing DDX3Y, a crucial autoimmunity gene, in hg-I and other common European Y-chromosome haplogroups looking for functional variants. We extended the genetic analyses to CD24L4 and compared and contrasted the roles of disease-based selection, demographic history and population structure shaping the contemporary genetic landscape of hg-I chromosomes. Our results confirmed and refined the AIDS progression signal to hg-I, though no gene variant was identified that can explain the disease association. Molecular evolutionary and genetic analyses of the examined loci suggested a unique evolutionary history in hg-I, probably shaped by complex interactions of selection, demographic history and high geographical differentiation leading to the formation of distinct hg-I subhaplogroups that today are associated with HIV/AIDS onset. Clearly, further studies on Y-chromosome candidate loci sequencing to discover functional variants and discern the roles of evolutionary factors are warranted.
June 24, 2010
Population structure in Ireland and Britain (O'Dushlaine et al. 2010)
 From the paper:
From the paper:Eigensoft PCA analysis across all seven of our European and European-ancestry populations broadly identified four sub-groups consisting of (i) Bulgarian, (ii) Portuguese, (iii) Swedish and (iv) Irish/British/Utah populations (see Figure 1a, Supplementary Figure S1). The first two principal components (PCs) separate out northern from southern, and western from eastern European ancestry, respectively. The Europe-wide PCA analysis positions the Scottish population (Aberdeen) intermediate between the Irish and English populations. We further explored this observation by restricting our PC analysis to residents of Ireland, Scotland (Aberdeen) and south/southeast England (Figure 1b, Supplementary Figure S1). This analysis confirms the observation that the Scottish population is intermediate between the Irish and English cohorts on the first principal component(this time dividing west from east). Although more subtle, the Scottish cohort is also shifted slightly from the other two on PC2.The distinction between Britons and Swedes was also noted in an earlier study. It's nice to see Bulgarians and Portuguese sampled, as they have been rather neglected in genomic studies, but, unfortunately none of their neighbors or any other intermediate populations were included, which is understandable as the study focused on British Isles populations. Bulgarians and Portuguese served as "anchor points" to re-create the well-known correlation of the first two PCs of European genetic variation with longitude/latitude.
In summary, our results illustrate a subtle genetic structure across Britain and Ireland in the context of the comparatively homogenous nature of the European genetic pool. We have observed slightly elevated levels of LD and genome-wide homozygosity in Ireland and Sweden compared with neighbouring British and European populations, although these levels do not approach those of traditional population isolates. Similarly, we have illustrated a decrease in HD in Britain and Ireland, more so in Scotland and Ireland than in England.

At K=2 we see a distinction between northern and southern Europeans.
Population structure and genome-wide patterns of variation in Ireland and Britain
Colm T O'Dushlaine et al.
Abstract
Located off the northwestern coast of the European mainland, Britain and Ireland were among the last regions of Europe to be colonized by modern humans after the last glacial maximum. Further, the geographical location of Britain, and in particular of Ireland, is such that the impact of historical migration has been minimal. Genetic diversity studies applying the Y chromosome and mitochondrial systems have indicated reduced diversity and an increased population structure across Britain and Ireland relative to the European mainland. Such characteristics would have implications for genetic mapping studies of complex disease. We set out to further our understanding of the genetic architecture of the region from the perspective of (i) population structure, (ii) linkage disequilibrium (LD), (iii) homozygosity and (iv) haplotype diversity (HD). Analysis was conducted on 3654 individuals from Ireland, Britain (with regional sampling in Scotland), Bulgaria, Portugal, Sweden and the Utah HapMap collection. Our results indicate a subtle but clear genetic structure across Britain and Ireland, although levels of structure were reduced in comparison with average cross-European structure. We observed slightly elevated levels of LD and homozygosity in the Irish population compared with neighbouring European populations. We also report on a cline of HD across Europe with greatest levels in southern populations and lowest levels in Ireland and Scotland. These results are consistent with our understanding of the population history of Europe and promote Ireland and Scotland as relatively homogenous resources for genetic mapping of rare variants.
Link
June 23, 2010
Genes predict village of origin in rural Europe (O'Dushlaine et al. 2010)
Using 300 000 SNPs and only unrelated, non-inbred individuals with all four grandparents from the same valley, village or isle, we here show the genomic differentiation across 8–30 km in three disparate areas of rural Europe, using genetic information alone (Figure 1). PCA of genomic sharing and model-based clustering (not shown) both allow separation of individuals with grandparents from each of three small Scottish isles, three alpine valleys in the north of Italy and two villages on one small island in Croatia. We used a supervised classification approach to predict subpopulation membership. Highly reliable levels of prediction were achieved with 100, 96 and 89% of individuals correctly classified on the basis of their genetic data for Italy, the Scottish Isles and Croatia, respectively.
European Journal of Human Genetics doi: 10.1038/ejhg.2010.92
Genes predict village of origin in rural Europe
Colm O'Dushlaine et al.
Abstract
Link
Modern humans bite hard
 From the paper:
From the paper:Our findings offer an explanation for the apparently inconsistent presence of a dentition in H. sapiens that appears well adapted to resist high bite forces relative to other extant hominids, set in a cranium and mandible that are relatively gracile and characterized by less robust musculature. Thus, the teeth of humans need to be able to resist comparable bite reaction forces to those of other extant hominids, but, because considerably less muscle force is required to achieve any given bite reaction force in the human than in other hominids, less stress is produced.Proceedings of the Royal Society B doi: 10.1098/rspb.2010.0509
...
We conclude that although humans are well adapted to produce high peak forces with the jaw moving in rotation, they may not be as well adapted to produce and maintain high bite forces with the jaw moving in translation. Thus, Homo sapiens may be comparable to other hominids in possessing an ability to access some relatively hard foods through the application of high transitory bite forces, however, our species may be less well adapted to consume tough or hard foods that require powerful, sustained chewing.
The craniomandibular mechanics of being human
Stephen Wroe et al.
Diminished bite force has been considered a defining feature of modern Homo sapiens, an interpretation inferred from the application of two-dimensional lever mechanics and the relative gracility of the human masticatory musculature and skull. This conclusion has various implications with regard to the evolution of human feeding behaviour. However, human dental anatomy suggests a capacity to withstand high loads and two-dimensional lever models greatly simplify muscle architecture, yielding less accurate results than three-dimensional modelling using multiple lines of action. Here, to our knowledge, in the most comprehensive three-dimensional finite element analysis performed to date for any taxon, we ask whether the traditional view that the bite of H. sapiens is weak and the skull too gracile to sustain high bite forces is supported. We further introduce a new method for reconstructing incomplete fossil material. Our findings show that the human masticatory apparatus is highly efficient, capable of producing a relatively powerful bite using low muscle forces. Thus, relative to other members of the superfamily Hominoidea, humans can achieve relatively high bite forces, while overall stresses are reduced. Our findings resolve apparently discordant lines of evidence, i.e. the presence of teeth well adapted to sustain high loads within a lightweight cranium and mandible.
Link
In search of Dionysos. Reassessing a Dionysian context in early Rome
In the present study the possibility of an early appearance of the god Dionysos and his sphere in archaic Rome, in the decades around 500 BC, will be examined.In early scholarship, rooted in the 19th century, the phenomenon of Dionysian ecstatic rites, cults, and satyr-plays in Roman society was denied. According to that view and the subsequent tradition in religious studies, such cultic activities were not present in Rome. Furthermore, due to Christian presuppositions, religion could scarcely be connected with sexual activities and bawdy behaviour, and as this is one fundamental quality in Dionysian cultic activities, it was reason enough for neglect and rejection of the thought of Dionysian cult as religion proper, on the whole. These preconceptions have long prevailed and formed the foundation for research in Roman religious studies. Scholars in various disciplines now challenge these ideas.The theoretical framework in this multidisciplinary study focuses on an intercontextual methodology and will have the approach of a case study. The starting point is thus to make a reassessment of the evidence at hand. The importance of the iconographic material is brought forward, beside the literary and epigraphic sources. Finds from the Greek and Etruscan areas supply a comparative perspective since Rome hardly can be seen as an isolated entity. It is suggested that ideas and values travelled rather freely in the area. Parallel Dionysian phenomena are known in the cultural spheres influencing Rome. Dionysos’ visual manifestations are in focus as well as Dionysos’ possible revelation in early Rome and plausible relation to the god Liber. Moreover, the diverse aspects of the satyrs as part of the Dionysian sphere are treated and an attempt is made to explain the satyr in a religious context. Liminality is a central feature when satyrs are concerned, and their function as a symbol of inversion of order is considered. Arguments are given for a strong connection between ritual and performance, thus indicating a cultic origin of performances in Rome, and for an early appearance of Dionysos and his thiasos.
June 21, 2010
Brown-eyed men perceived to be more dominant

The question arises: why are brown-eyed males rated as more dominant than blue-eyed? Some facial features such as square jaws, thick eyebrows and broad cheekbones are linked with higher perceived dominance; facial submissiveness, on the other hand, is characterized by a round face with large eyes, smallish nose, and high eyebrows (Berry, 1990; Berry & Mcarthur, 1986; Cunningham,Barbee, & Pike, 1990; Mazur, Halpern, & Udry, 1994; Mueller & Mazur,1997; Thornhill & Gangestad, 1994). The morphological differences between blue-eyed and brown-eyed males were visualized by deformation of thin-plate splines (Fig. 3). In contrast with blue-eyed males, brown-eyed males have statistically broader and rather massive chins, broader (laterally prolonged) mouths, larger noses, and eyes that are closer together with larger eyebrows. In contrast, blue-eyed males show smaller and sharper chins, mouths that are laterally narrower, noses smaller, and a greater span between the eyes. Especially the broader massive chin, bigger nose, and larger eyebrows of brown-eyed males may explain their higher perceived dominance.
The authors propose that true genetic linkage between eye color and these other facial features is unlikely, as eye color is determined by few loci, and these are unlikely to be the same ones that influence these other facial features. Thus, they propose a different explanation, namely that blue-eyed and brown-eyed children are treated differently by their parents as they grow up, and this "different treatment" manifests itself phenotypically. The argument in favor of different treatment stems from the fact that many people are born blue-eyed, but their eye color is set to a darker shade eventually. The authors write:
It is possible that subjects with blue eyes are treated as a small child for a longer period than brown-eyed children. Such early social experience may have been literally ‘‘inscribed” into their faces, preserved until adulthood, and finally bring on the perception of higher submissiveness. Rosenberg and Kagan (1987, 1989) investigated the association between eye color and behavioral inhibition, revealing that children with blue eyes are more inhibited. Coplan et al. (1998) found a significant interaction between eye color and social wariness within preschoolers. Blueeyed males were rated as more socially wary, i.e. being more temperamentally inhibited, displaying more reticent behavior and having more internalizing problems, than males with brown eyes, though there were no differences between blue- and brown-eyed females (Coplan et al., 1998).
There is an alternative explanation, that requires neither genetic linkage nor an environmental factor such as upbringing. That factor is latent population structure.
In a truly long-term random mating population, and assuming that eye color is not genetically linked with e.g. chin breadth, then all combinations of chin breadth and eye color would occur with a probability determined entirely by the frequency of their genetic determinants in the population.
However, consider the possibility that the population is an incomplete mixture of a blue-eyed "facially submissive" population element, and a brown-eyed "facially dominant" one. If that was the case, then brown-eyed folks would tend to have dominant facial features by reason of their ancestry rather than any genetic linkage between the two traits.
[1] International anthropometric study of facial morphology in various ethnic groups/races, Leslie G Farkas, J Craniofac Surg 16:615-46
Personality and Individual Differences
Volume 49, Issue 1, July 2010, Pages 59-64
Eye color predicts but does not directly influence perceived dominance in men
Karel Kleisner et al.
This study focuses on the relationship between eye color, gender, and psychological characteristics perceived from the human face. Photographs of 40 male and 40 female students were rated for perceived dominance and attractiveness. Attractiveness showed no relation with eye color. In contrast, eye color had a significant effect on perceived dominance in males: brown-eyed men were rated as more dominant than men with blue eyes. To control for the effect of eye color, we studied perceived dominance on the same photographs of models after changing the iris color. The eye color had no effect on perceived dominance. This suggests that some other facial features associated with eye color affect the perception of dominance in males. Geometric morphometrics have been applied to reveal features responsible for the differences in facial morphospace of blue-eyed and brown-eyed males.
Link
June 20, 2010
Radiocarbon based chronology for ancient Egypt (Ramsey et al. 2010)
DOI: 10.1126/science.1189395
Radiocarbon-Based Chronology for Dynastic Egypt
Christopher Bronk Ramsey et al.
The historical chronologies for dynastic Egypt are based on reign lengths inferred from written and archaeological evidence. These floating chronologies are linked to the absolute calendar by a few ancient astronomical observations, which remain a source of debate. We used 211 radiocarbon measurements made on samples from short-lived plants, together with a Bayesian model incorporating historical information on reign lengths, to produce a chronology for dynastic Egypt. A small offset (19 radiocarbon years older) in radiocarbon levels in the Nile Valley is probably a growing-season effect. Our radiocarbon data indicate that the New Kingdom started between 1570 and 1544 B.C.E., and the reign of Djoser in the Old Kingdom started between 2691 and 2625 B.C.E.; both cases are earlier than some previous historical estimates.
Link
June 19, 2010
Wealth and people and the invasion of alien species
PNAS doi:10.1073/pnas.1002314107
Disentangling the role of environmental and human pressures on biological invasions across Europe
Petr Pyšek et al.
The accelerating rates of international trade, travel, and transport in the latter half of the twentieth century have led to the progressive mixing of biota from across the world and the number of species introduced to new regions continues to increase. The importance of biogeographic, climatic, economic, and demographic factors as drivers of this trend is increasingly being realized but as yet there is no consensus regarding their relative importance. Whereas little may be done to mitigate the effects of geography and climate on invasions, a wider range of options may exist to moderate the impacts of economic and demographic drivers. Here we use the most recent data available from Europe to partition between macroecological, economic, and demographic variables the variation in alien species richness of bryophytes, fungi, vascular plants, terrestrial insects, aquatic invertebrates, fish, amphibians, reptiles, birds, and mammals. Only national wealth and human population density were statistically significant predictors in the majority of models when analyzed jointly with climate, geography, and land cover. The economic and demographic variables reflect the intensity of human activities and integrate the effect of factors that directly determine the outcome of invasion such as propagule pressure, pathways of introduction, eutrophication, and the intensity of anthropogenic disturbance. The strong influence of economic and demographic variables on the levels of invasion by alien species demonstrates that future solutions to the problem of biological invasions at a national scale lie in mitigating the negative environmental consequences of human activities that generate wealth and by promoting more sustainable population growth.
Link
June 18, 2010
Mediterranean diet improves autonomic heart function among middle-aged men
Mediterranean Dietary Pattern Is Associated With Improved Cardiac Autonomic Function Among Middle-Aged Men
A Twin Study
Jun Dai et al.
Background Reduced heart rate variability (HRV), a measure of cardiac autonomic dysfunction, is a risk factor for coronary artery disease. Diet can influence HRV, but this association may be confounded by genetic and environmental factors.
Methods and Results We administered the Willett Food Frequency Questionnaire to 276 middle-aged male twins. We derived a score measuring the extent to which an individual's diet conformed to the Mediterranean diet following a published algorithm. The higher the score, the greater the similarity to the Mediterranean diet. All twins underwent 24-hour ambulatory ECG recording. Time and frequency domain measures of HRV were calculated. Mixed-effects regression was used to partition the association into between- and within-twin pair differences. After adjusting for energy intake, other nutritional factors, shared genes, and common environment, a 1-unit higher score was significantly associated with 3.9% to 13% higher time and frequency domain HRV parameters. Further controlling for known cardiovascular risk factors and use of fish oil supplements and medications did not substantially change the estimates.
Conclusions The Mediterranean dietary pattern is associated with higher HRV.
Link
June 17, 2010
Y-chromosome of Caravaggio
 From the Guardian:
From the Guardian:Scientists seeking to shed light on the mysterious death of the Italian artist in 1610 said they are "85% sure" they have found his bones thanks to carbon dating and DNA checks on remains excavated in Tuscany.A poster at dna-forums submitted this screenshot of the Merisi results:
...
Michelangelo Merisi, known as Caravaggio after the Lombardy town where he grew up, was a young man at the height of his career in Rome when he killed a man in a brawl in 1606, fleeing to find new patrons in Naples and then Malta, only to be thrown off the island two years later for more brawling.
...
The team's next stop was the town of Caravaggio to compare DNA from the bones with local people. No descendents were found but families with the same surname were traced, giving samples which were 50 to 60% compatible with the bones.

Autosomal haplotype shared by Arab Muslims and Oriental Jews
An ancient autosomal haplotype bearing a rare achromatopsia-causing founder mutation is shared among Arab Muslims and Oriental Jews
Lina Zelinger et al.
Numerous cultural aspects, mainly based on historical records, suggest a common origin of the Middle-Eastern Arab Muslim and Jewish populations. This is supported, to some extent, by Y-chromosome haplogroup analysis of Middle-Eastern and European samples. Up to date, no genomic regions that are shared among Arab Muslim and Jewish chromosomes and are unique to these populations have been reported. Here, we report of a rare achromatopsia-causing CNGA3 mutation (c.1585G>A) presents in both Arab Muslim and Oriental Jewish patients. A haplotype analysis of c.1585G>A-bearing chromosomes from Middle Eastern and European origins revealed a shared Muslim–Jewish haplotype, which is different from those detected in European patients, indicating a recurrent mutation stratified by a Jewish–Muslim founder effect. Comprehensive whole-genome haplotype analysis using 250 K single nucleotide polymorphism arrays revealed a large homozygous region of ~11 Mbp shared by both Arab Muslim and Oriental Jewish chromosomes. A subsequent microsatellite analysis of a 21.5 cM interval including CNGA3 and the adjacent chromosome 2 centromere revealed a unique and extremely rare haplotype associated with the c.1585G>A mutation. The age of the shared c.1585G>A mutation was calculated using the microsatellite genotyping data to be about 200 generations ago. A similar analysis of mutation age based on the Arab Muslim data alone showed that the mutation was unlikely to be the product of a recent gene flow event. The data present here demonstrate a large (11 Mbp) genomic region that is likely to originate from an ancient common ancestor of Middle-Eastern Arab Muslims and Jews who lived approximately 5,000 years ago.
Link
June 16, 2010
Two waves of expansion from East Asia into the Americas
 Related:
Related:- Concurrent but distinctive migrations into the Americas
- Settlement of Americas: one migration with recurrent gene flow
Testing Evolutionary and Dispersion Scenarios for the Settlement of the New World
Mark Hubbe et al.
Abstract
Background
Discussion surrounding the settlement of the New World has recently gained momentum with advances in molecular biology, archaeology and bioanthropology. Recent evidence from these diverse fields is found to support different colonization scenarios. The currently available genetic evidence suggests a “single migration” model, in which both early and later Native American groups derive from one expansion event into the continent. In contrast, the pronounced anatomical differences between early and late Native American populations have led others to propose more complex scenarios, involving separate colonization events of the New World and a distinct origin for these groups.
Methodology/Principal Findings
Using large samples of Early American crania, we: 1) calculated the rate of morphological differentiation between Early and Late American samples under three different time divergence assumptions, and compared our findings to the predicted morphological differentiation under neutral conditions in each case; and 2) further tested three dispersal scenarios for the colonization of the New World by comparing the morphological distances among early and late Amerindians, East Asians, Australo-Melanesians and early modern humans from Asia to geographical distances associated with each dispersion model. Results indicate that the assumption of a last shared common ancestor outside the continent better explains the observed morphological differences between early and late American groups. This result is corroborated by our finding that a model comprising two Asian waves of migration coming through Bering into the Americas fits the cranial anatomical evidence best, especially when the effects of diversifying selection to climate are taken into account.
Conclusions
We conclude that the morphological diversity documented through time in the New World is best accounted for by a model postulating two waves of human expansion into the continent originating in East Asia and entering through Beringia.
June 15, 2010
June 13, 2010
Composite Greek player (FIFA 2010 World Cup)

IFIH1 points to ancient structure in Africa
Population genetics of IFIH1: ancient population structure, local selection and implications for susceptibility to type 1 diabetes
Matteo Fumagalli et al.
The human IFIH1 gene encodes a sensor of double strand RNA involved in innate immunity against viruses, indicating that this gene is a likely target of virus-driven selective pressure. Notably, IFIH1 also plays a role in autoimmunity as common and rare polymorphisms in this gene have been associated with type 1 diabetes (T1D). We analysed the evolutionary history of IFIH1 in human populations. Results herein suggest that two major IFIH1 haplotype clades originated from ancestral population structure (or balancing selection) in the African continent and that local selective pressures have acted on the gene. Specifically, directional selection in Europe and Asia resulted in the spread of a common IFIH1 haplotype carrying a derived His460 allele. This variant changes a highly conserved arginine residue in the helicase domain, possibly conferring altered specificity in viral recognition. An alternative common haplotype has swept to high frequency in South Americans as a result of recent positive selection. Previous studies suggested that a portion of risk alleles for autoimmune diseases could have been maintained in humans as they conferred a selective advantage against infections. This is not the case for IFIH1 as population genetic differentiation and haplotype analyses indicated that the T1D susceptibility alleles behaved as neutral or nearly neutral polymorphisms. Our findings suggest that variants in IFIH1 confer different susceptibility to diverse viral infections and provide insight into the relationship between adaptation to past infection and predisposition to autoimmunity in modern populations.
Link
June 12, 2010
Composite faces of Iran
June 09, 2010
Genome-wide structure of Jews (Behar et al. 2010)
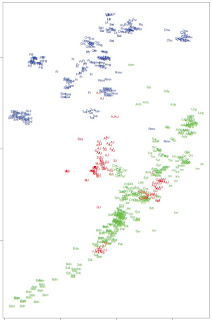 On the left, PCA from Supplementary Figure 3, shows clearly at least three different Jewish clusters. Note the main Ashkenazi/Sephardi cluster halfway between Tuscans and Near Eastern populations, a Yemeni Jewish cluster coinciding with Bedouins and Saudi Arabians, and a West Asian cluster encompassing Georgian, Iranian, Iraqi, etc. Jews.
On the left, PCA from Supplementary Figure 3, shows clearly at least three different Jewish clusters. Note the main Ashkenazi/Sephardi cluster halfway between Tuscans and Near Eastern populations, a Yemeni Jewish cluster coinciding with Bedouins and Saudi Arabians, and a West Asian cluster encompassing Georgian, Iranian, Iraqi, etc. Jews.Below is ADMIXTURE analysis in the global context.
There is a ton of information in the above figure, for Jews and non-Jews alike. Some observations:
- Ethiopians and Ethiopian Jews look identical, between Sub-Saharan Africans and West Asians .
- Sub-Saharan admixture in Egyptians and Yemenites is quite evident; lack of such admixture in Europe and non-Arabs from West Asia.
- A little Caucasoid admixture in Mongols
- Split of Mongoloids into two clusters, which appear to be "northern" and "southern"
- Central Asian Turkic speakers (Uygur, Uzbek) derived from both Mongoloid sub-clusters; their Caucasoid components are mainly West Asian (light green) rather than north European (dark blue)
- Non-European components in Russians are resolved into Caucasoid light green and "north Mongoloid" (see above)
- A little of the "north Mongoloid" component in Turks and some populations from the Caucasus, not much elsewhere in West Eurasia
- South Asian (green) component in Cambodians
- Russians and Lithuanians lack south European (light blue) component but have some west Asian (light green)
- Cypriots are split between West Asia and Southern European components, with minority Semitic (Phoenicians or Syrian Christians?) and northern European ones.
- French Basque and Sardinians lack West Asian component (light green)
This study further uncovers genetic structure that partitions most Jewish samples into Ashkenazi–north African– Sephardi, Caucasus–Middle Eastern, and Yemenite subclusters (Fig. 2). There are several mutually compatible explanations for the observed pattern: a splintering of Jewish populations in the early Diaspora period, an underappreciated level of contact between members of each of these subclusters, and low levels of admixture with Diaspora host populations.
The genome-wide structure of the Jewish people
Doron M. Behar et al.
Contemporary Jews comprise an aggregate of ethno-religious communities whose worldwide members identify with each other through various shared religious, historical and cultural traditions1, 2. Historical evidence suggests common origins in the Middle East, followed by migrations leading to the establishment of communities of Jews in Europe, Africa and Asia, in what is termed the Jewish Diaspora3, 4, 5. This complex demographic history imposes special challenges in attempting to address the genetic structure of the Jewish people6. Although many genetic studies have shed light on Jewish origins and on diseases prevalent among Jewish communities, including studies focusing on uniparentally and biparentally inherited markers7, 8, 9,10, 11, 12, 13, 14, 15, 16, genome-wide patterns of variation across the vast geographic span of Jewish Diaspora communities and their respective neighbours have yet to be addressed. Here we use high-density bead arrays to genotype individuals from 14 Jewish Diaspora communities and compare these patterns of genome-wide diversity with those from 69 Old World non-Jewish populations, of which 25 have not previously been reported. These samples were carefully chosen to provide comprehensive comparisons between Jewish and non-Jewish populations in the Diaspora, as well as with non-Jewish populations from the Middle East and north Africa. Principal component and structure-like analyses identify previously unrecognized genetic substructure within the Middle East. Most Jewish samples form a remarkably tight subcluster that overlies Druze and Cypriot samples but not samples from other Levantine populations or paired Diaspora host populations. In contrast, Ethiopian Jews (Beta Israel) and Indian Jews (Bene Israel and Cochini) cluster with neighbouring autochthonous populations in Ethiopia and western India, respectively, despite a clear paternal link between the Bene Israel and the Levant. These results cast light on the variegated genetic architecture of the Middle East, and trace the origins of most Jewish Diaspora communities to the Levant.
Link
June 08, 2010
Guess the origin of this composite man
Osteogenin (BMP3) selection in humans
 On the left: Figure 5. Worldwide distribution of the allele frequency of the SNP rs3733549 (Arg192Gln) genotyped in this study.
On the left: Figure 5. Worldwide distribution of the allele frequency of the SNP rs3733549 (Arg192Gln) genotyped in this study.From the paper:
There is a high degree of divergence of haplotype composition between African and Non-Africans. Two major clades account for ~78.3% haplotypes for CEU, and these are also the major haplotypes in East Asians (HCB+JPT) but are rare (only ~5.3%) in Africans (YRI).
...
The adaptive evolution of BMP3 is consistent with the rapid evolution of the human skeletal system, although we do not have data that explains the mechanism for the selective advantage of the BMP3 variant. BMP3, is an antagonist of several osteogenic BMPs, and is a negative determinant of bone density [19]. Lacking the BMP3, mice have increased bone mass [19]. Potentially, the antagonistic activity of human BMP3 to osteogenic acting factors, and even the level of BMP signal, was adaptively changed via many amino acid substitutions during human evolution, which may diverge functionally from chimpanzee accounting for the skeletal differences [2], [30]. It still needs test by further functional experiment. The targets of selection operated on the BMP3 are different between European and East Asian evidenced by long-range haplotype test (Fig. 2). Within modern human populations, BMP3 may also diverge in the activity, expression level, accounting for the skeletal variation, such as body mass, because of the key function of BMP3 in the skeletal system [19], [31], [32].
PLoS ONE doi:10.1371/journal.pone.0010959
Evidence for Positive Selection on the Osteogenin (BMP3) Gene in Human Populations
Dong-Dong Wu et al.
Abstract
Background
Human skeletal system has evolved rapidly since the dispersal of modern humans from Africa, potentially driven by selection and adaptation. Osteogenin (BMP3) plays an important role in skeletal development and bone osteogenesis as an antagonist of the osteogenic bone morphogenetic proteins, and negatively regulates bone mineral density.
Methodology/Principal Findings
Here, we resequenced the BMP3 gene from individuals in four geographically separated modern human populations. Features supportive of positive selection in the BMP3 gene were found including the presence of an excess of nonsynonymous mutations in modern humans, and a significantly lower genetic diversity that deviates from neutrality. The prevalent haplotypes of the first exon region in Europeans demonstrated features of long-range haplotype homogeneity. In contrast with findings in European, the derived allele SNP Arg192Gln shows higher extended haplotype homozygosity in East Asian. The worldwide allele frequency distribution of SNP shows not only a high-derived allele frequency in Asians, but also in Americans, which is suggestive of functional adaptation.
Conclusions/Significance
In conclusion, we provide evidence for recent positive selection operating upon a crucial gene in skeletal development, which may provide new insight into the evolution of the skeletal system and bone development.
June 07, 2010
Facial bone structure changes with age
Regional shape change in adult facial bone curvature with age
Shanna E. Williams, Dennis E. Slice
Life expectancies have increased dramatically over the last 100 years, affording greater opportunities to study the impact of age on adult craniofacial morphology. This article employs a novel application of established geometric morphometric methods to examine shape differences in adult regional facial bone curvature with age. Three-dimensional semilandmarks representing the curvature of the orbits, zygomatic arches, nasal aperture, and maxillary alveolar process were collected from a cross-sectional cranial sample of mixed sex and ancestry (male and female; African- and European-American), partitioned into three age groups (young adult = 18-39; middle-aged = 40-59 years; and elderly = 60+ years). Each facial region's semilandmarks were aligned into a common coordinate system via generalized Procrustes superimposition. Regional variation in shape was then explored via a battery of multivariate statistical techniques. Age-related shape differences were detected in the orbits, zygomatic arches, and maxillary alveolar process. Interactions between age, sex, and ancestry were also identified. Vector plots revealed patterns of superoinferior compression, lateral expansion, and posterior recession depending on the population/subpopulation, location, and age groups examined. These findings indicate that adult craniofacial curvature shape is not static throughout human life. Instead, age-related spatial modifications occur in various regions of the craniofacial skeleton. Moreover, these regional alterations vary not only through time, but across human populations and the sexes.
Link
June 03, 2010
Two major groups of living Jews (Atzmon et al. 2010)
Next, each of 2407 European subjects was assigned into one of 10 groups based on geographic region: South:Italy, Swiss-Italian; Southeast: Albania, Bosnia-Herzegovina, Bulgaria, Croatia, Greece, Kosovo, Macedonia, Romania, Serbia,Slovenia, Yugoslavia; Southwest: Portugal, Spain; East: CzechRepublic, Hungary; East-Southeast: Cyprus, Turkey; Central:Austria, Germany, Netherlands, Swiss-German; West: Belgium,France, Swiss-French, Switzerland; North: Denmark, Norway,Sweden; Northeast: Finland, Latvia, Poland, Russia, Ukraine;Northwest: Ireland, Scotland, UK.


Admixture with local populations, including Khazars and Slavs, may have occurred subsequently during the 1000 year (2nd millennium) history of the European Jews. Based on analysis of Y chromosomal polymorphisms, Hammer estimated that the rate might have been as high as 0.5% per generation or 12.5% cumulatively (a figure derived from Motulsky), although this calculation might have underestimated the influx of European Y chromosomes during the initial formation of European Jewry. Notably, up to 50% of Ashkenazi Jewish Y chromosomal haplogroups (E3b, G, J1, and Q) are of Middle Eastern origin,15 whereas the other prevalent haplogroups (J2, R1a1, R1b) may be representative of the early European admixture. The 7.5% prevalence of the R1a1 haplogroup among Ashkenazi Jews has been interpreted as a possible marker for Slavic or Khazar admixture because this haplogroup is very common among Ukrainians (where it was thought to have originated), Russians, and Sorbs, as well as among Central Asian populations, although the admixture may have occurred with Ukrainians, Poles, or Russians, rather than Khazars. In support of the ancestry observations reported in the current study, the major distinguishing feature between Ashkenazi and Middle Eastern Jewish Y chromosomes was the absence of European haplogroups in Middle Eastern Jewish populations.
AJHG doi:10.1016/j.ajhg.2010.04.015
Abraham's Children in the Genome Era: Major Jewish Diaspora Populations Comprise Distinct Genetic Clusters with Shared Middle Eastern Ancestry
Gil Atzmon et al.
Abstract
For more than a century, Jews and non-Jews alike have tried to define the relatedness of contemporary Jewish people. Previous genetic studies of blood group and serum markers suggested that Jewish groups had Middle Eastern origin with greater genetic similarity between paired Jewish populations. However, these and successor studies of monoallelic Y chromosomal and mitochondrial genetic markers did not resolve the issues of within and between-group Jewish genetic identity. Here, genome-wide analysis of seven Jewish groups (Iranian, Iraqi, Syrian, Italian, Turkish, Greek, and Ashkenazi) and comparison with non-Jewish groups demonstrated distinctive Jewish population clusters, each with shared Middle Eastern ancestry, proximity to contemporary Middle Eastern populations, and variable degrees of European and North African admixture. Two major groups were identified by principal component, phylogenetic, and identity by descent (IBD) analysis: Middle Eastern Jews and European/Syrian Jews. The IBD segment sharing and the proximity of European Jews to each other and to southern European populations suggested similar origins for European Jewry and refuted large-scale genetic contributions of Central and Eastern European and Slavic populations to the formation of Ashkenazi Jewry. Rapid decay of IBD in Ashkenazi Jewish genomes was consistent with a severe bottleneck followed by large expansion, such as occurred with the so-called demographic miracle of population expansion from 50,000 people at the beginning of the 15th century to 5,000,000 people at the beginning of the 19th century. Thus, this study demonstrates that European/Syrian and Middle Eastern Jews represent a series of geographical isolates or clusters woven together by shared IBD genetic threads.