This is a quite interesting paper, as it shows, by sampling a large number of individuals), that the heritability of height is not missing after all. The authors looked at a large number of individuals, and this allowed them to discover statitically significant associations between height and more SNPs than before.
This bears great promise as it may hint that genome-wide association studies, that have come under substantial criticism lately, may be failing not because of an inherent flaw, but rather because they are not sampling enough individuals.
The discovered SNPs account for 45% of the heritability of height. Where is the rest? The authors argue for two additional sources:
First, SNPs in current microarray chips sample the genome incompletely. Locations in-between discovered SNPs are in incomplete linkage disequilibrium with the discovered SNPs. So, there is undetected polymorphism, in the gaps between the hundreds of thousands of SNPs in current chips, that may explain a portion of the missing heritability.
Second, SNPs have different minor allele frequencies. For example, in one SNP the minor allele may occur at 10% of individuals, while in others at 30%. This is important, because it is more difficult to arrive at a statistically significant result in the former case.
Consider a SNP with a minor allele frequency of 2%. Then, if you sample 1,000 individuals, only about 20 of them are expected to have the minor allele. You cannot estimate the average height of the minor allele with a sample of 20 people as securely as you can with a sample of 500. Thus, if the SNP influences height in a small way, you will not be able to detect it.
A further complication, which I've written about before, is that some variation in the human genome is family-related, or at least occurs at fewer individuals than the allele frequency cutoff. If 99.9% of people have C at a given location and 0.1% of people have T, this variant is unlikely to be included in a microarray chp, because it is too rare to matter economically: you would only get a handful of individuals -if you're lucky- in a sample of 1,000 for such a variant. However, rarity does not mean that the variant is functionally unimportant, and the rare allele may play a substantial role in the height of the people who possess it.
The publication of this paper is a cause for optimism, as it shows that progress can be made by brute force: fuller genome coverage and more individuals. We'll have to wait and see whether or not the same approach will work for other complex traits, such as IQ or schizophrenia, that have been hitherto difficult to crack.
Obviously, the cost of sampling more individuals will become an issue in future studies, but the cost-per-individual is expected to drop. So, I'm guessing that more discoveries are in store for us in the next few years.
UPDATE (Jun 28):
Not the main point of the paper, but also included in the supplementary material (pdf) are some nice PCA results.
In the European-only PCA we see the familiar north-south gradient (anchored by Tuscans TSI and Netherlands NET on either side), and the orthogonal deviation of the Finns. Swedes (SWE) occupy a northern European end of the spectrum like the Dutch, but are spread towards Finns, reflecting low-level Finnish admixture in that population. Conversely, Finns are variable along the same axis, reflecting variable levels of admixture. Australians (AUS) and UK, on the other hand, are on the northern European edge of the main European gradient, with a number of individuals spread toward the Tuscan side.
In the European-only PCA we see the familiar north-south gradient (anchored by Tuscans TSI and Netherlands NET on either side), and the orthogonal deviation of the Finns. Swedes (SWE) occupy a northern European end of the spectrum like the Dutch, but are spread towards Finns, reflecting low-level Finnish admixture in that population. Conversely, Finns are variable along the same axis, reflecting variable levels of admixture. Australians (AUS) and UK, on the other hand, are on the northern European edge of the main European gradient, with a number of individuals spread toward the Tuscan side.
 The PCA with all populations is also quite interesting. East Eurasians (Chinese and Japanese) form a tight pole at the bottom right. Gujarati Indians (GIH) form a different pole, spread towards Europeans, reflecting variable levels of West Eurasian admixture in that population, probably corresponding to the ANI element recently discovered in Indian populations. Mexicans (MEX) are spread towards East Asians, reflecting their Amerindian admixture, but notice how they are not positioned exactly on the European-East Asian axis, probably reflecting the third, minority, Sub-Saharan element in their ancestry, as well as the fact that Amerindians are not perfectly represented by East Asians. Finns are tilted towards East Asians, as expected, reflecting the fact that their genetic specificity vis a vis Northern Europeans is due to low-level East Eurasian ancestry.
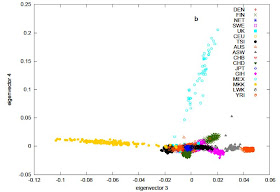
The PCA with all populations is also quite interesting. East Eurasians (Chinese and Japanese) form a tight pole at the bottom right. Gujarati Indians (GIH) form a different pole, spread towards Europeans, reflecting variable levels of West Eurasian admixture in that population, probably corresponding to the ANI element recently discovered in Indian populations. Mexicans (MEX) are spread towards East Asians, reflecting their Amerindian admixture, but notice how they are not positioned exactly on the European-East Asian axis, probably reflecting the third, minority, Sub-Saharan element in their ancestry, as well as the fact that Amerindians are not perfectly represented by East Asians. Finns are tilted towards East Asians, as expected, reflecting the fact that their genetic specificity vis a vis Northern Europeans is due to low-level East Eurasian ancestry. An interesting aspect of the first two PCs is the fact that the Maasai (MKK) and Luhya (LUW) from Kenya are not separated from Caucasoids, and neither are Yoruba from Nigeria (YRI). This is a good reminder of the fact that identity in the first two principal components may mask difference revealed in higher order components. This difference (at least for Maasai) is seen in the next two PCs.
An interesting aspect of the first two PCs is the fact that the Maasai (MKK) and Luhya (LUW) from Kenya are not separated from Caucasoids, and neither are Yoruba from Nigeria (YRI). This is a good reminder of the fact that identity in the first two principal components may mask difference revealed in higher order components. This difference (at least for Maasai) is seen in the next two PCs.Common SNPs explain a large proportion of the heritability for human height
Jian Yang et al.
Abstract
SNPs discovered by genome-wide association studies (GWASs) account for only a small fraction of the genetic variation of complex traits in human populations. Where is the remaining heritability? We estimated the proportion of variance for human height explained by 294,831 SNPs genotyped on 3,925 unrelated individuals using a linear model analysis, and validated the estimation method with simulations based on the observed genotype data. We show that 45% of variance can be explained by considering all SNPs simultaneously. Thus, most of the heritability is not missing but has not previously been detected because the individual effects are too small to pass stringent significance tests. We provide evidence that the remaining heritability is due to incomplete linkage disequilibrium between causal variants and genotyped SNPs, exacerbated by causal variants having lower minor allele frequency than the SNPs explored to date.
Link

Hi, Dienekes. Interesting; both the paper (for what I could read) and your analysis.
ReplyDeleteI imagine however that it will be most difficult to do GWAS when many or most traits' genetics are distributed probably in so many small subsets of people, right? It would seem (on first sight at least) that many traits are not defined by single instances of widespread genes but by a wide variety of roughly synonymous but different genes, right?
...
On a separate matter and rather a minor issue, I'd like if someone could clarify my perplexity at supp. fig. 2a (the supplementary material is freely available, it seems).
The legend "explains" that the graphs are "principal component analysis (PCA) of ancestry", where the Australian sample (and only them) was not used to generate the PC space but introduced on it after that. And that for fig. 2a: "The major trend, Principal Component (eigenvector, PC) 1, tends to separate African from non-African population while PC2 separate East Asian from the others".
That would be what one could expect (based on all other data I know) but the actual plot shows that PC1 separates East Asians from the rest and PC2 Indians (GIH) from the rest, with Europeans and African clumping together in the same corner.
I'm totally amiss if this is an error or what and would love if someone could explain.
There are three different African populations, so I tend to think that the figure shows what it shows, i.e., Gujarati Indians on the top.
ReplyDeleteAfricans are indeed cleanly separated from non-Africans on PC1, however, it is just that the there is a small visible gap between MKK/YRI/LWK and Europeans.
"... it is just that the there is a small visible gap between MKK/YRI/LWK and Europeans".
ReplyDeletePrecisely: in practical terms Europeans and Africans cluster together. This is totally contradictory with what we could expect (based on all other available data) and with the legend. I suspect it must be an error, with the wrong graphs being plotted instead of the correct ones. Otherwise it doesn't make any sense.
Unfortunate that they didn't sample people of the Dinaric Alps.
ReplyDeleteThe legend "explains" that the graphs are "principal component analysis (PCA) of ancestry", where the Australian sample (and only them) was not used to generate the PC space but introduced on it after that.
ReplyDeleteBecause a minority of Australians tested had been excluded from the PCA plots due to their obvious non-European ancestry, so using the rest of Australians (however bulk of them) in the generation of the PCs would give an unnaturalness (however little) to the PCA plots, as that would affect the non-Australian populations included in the PCA plots.
Good point Maju
ReplyDeleteIt looks to me like the Eigenvector 2 scale for the Gujarati is different from the rest. I expected it to track along the line towards the Asians.
Plus the range on the Eigenvector 2scale is much too large for the rest of the populations, scrunching up the data vertically.
I realized the reason of the confusion about the PC plots. Well, the PC plots should be viewed as a 4 sided pyramid (tetrahedron), with 4 vortices. Try to visualize that. That's why Africans and European look really close, but they're only close bidimensionally. If viewed 3D, one wold notice they are just 2 of the 4 vortices of an imaginary tetrahedron. For easy visualization: http://en.wikipedia.org/wiki/Tetrahedron
ReplyDeleteRangel-Villalobos H et al 2008, as there's a difference between Western and Eastern Mexicans, with a % of African ancestory between both.
ReplyDelete@pp987:You just made that up (or in any case it's clearly wrong).
ReplyDeleteThe PC space is a bidimensional euclidean space. In a few cases, where the PC3 axis is included in form of color or a projection of of a 3D space, then you'd be partly right (it's be actually an octahedron or cube, depending on what do you emphasize) but it's not the case here.
It must be an error where a different plot has been printed instead of the one that should be there. It has no other explanation.
Said that I wouldn't mind to know what that graph actually refers to (maybe PC3-PC4?) because it does indicate an Euro-African affinity at some level that is not so surprising to me. But cannot be at PC1-2 level because we all have seen by now dozens of such global PC1-2 plots and they do not produce these results.
Given the strong association of diet and stature, I'd be suspicious in an international sample of assuming that correlations between SNPs and stature are really genetic causes.
ReplyDeleteAncestry specific traits that happen to coincide with diet may look experimentally like a source of heritability of height.
I see no reason to think that the PC plot is wrong. People forget that the axes of PCA are not set in stone but are a function of the individuals included.
ReplyDeleteSure, globally, the greater genetic distinction is between Sub-Saharan Africans and Eurasians, but you are not guaranteed to get that every time you run PCA on a dataset.
To give an intuitive example, imagine having to explain variation with a single variable (i.e. PC1) in a room with 100 Japanese, 100 Englishmen, and 10 Nigerians. Nigerians are about 50% more distant from Englishmen than Japanese are, but there are 10 times more Japanese in the room than Nigerians. So, if you used PC1 to capture the Sub-Saharan/Eurasian distinction you would not capture most of the variation that could be captured with a single variable.
Going back to the figure, I see at least two reasons why Europeans and Africans are close in the first two PCs.
First of all, this plot, unlike previous ones many people are familiar with, is heavy on the East Africans. East Africans occupy an intermediate position between Yoruba and Europeans, and are thus as close to them (in a rough sense) as Mongoloids are.
Second, the plot includes both Gujarati Indians, and Mexicans. Thus, you already have two populations that don't map well in a 3-pole scheme (Africans, East Asians, Europeans) as the former are Caucasoid-South Asian and the latter are European-Amerindian.
In short there is no a priori reason to think that the plot is flawed. It could be, of course, but there's no rule set in stone that PCA will always produce a 3-pole African/European/East Asian in its first two components.
Not sure if what you say, Dienekes, is correct or not. In theory it could make some sense but in practice the plot is clearly contradicted by the legend:
ReplyDelete"The major trend, Principal Component (eigenvector, PC) 1, tends to separate African from non-African population" (no, that's not what it does: it separates East Asians from the rest) "... while PC2 separate East Asian from the others" (no, that's PC1, PC 2 in that plot separates Indians from the rest).
It happens the same with the PC3-4 graph: no correlation with the description at the legend.
It's a clear error, sorry for raising the issue.
I did say that the plot COULD be wrong.What the mixup is about remains to be seen (whether they put the wrong plot, or were sloppy with their description).
ReplyDeleteAnother good example of what I am referring to is from the recent study on Qatar where the first principal component is the Asian one, while the Sub-Saharan one is on the second component. In general, even though the greatest contrast within the human species is between Sub-Saharan Africans and Eurasians, it is not always guaranteed that this contrast will be mapped on the first PC.
I totally agree that PCA, like other statistical measures, is indeed subject to distortions because of sampling bias and/or subtle factors.
ReplyDeleteThe case of Qatar is probably caused by the fact that Qataris, like other peninsular Arabs, have some meaningful ultra-Saharan ancestry, what makes them closer overall to Africans than to East Asians. In this case African ancestry of Qatari is the "subtle factor" and the emphasis on Qataris the "sample bias", intentional in that study.
But, even then, the Global PC graph clearly shows the triangular structure that we are used to. And South Asians are represented there too, as is obvious from the scatter of the "Asian" sample between West Eurasia and East Asia. The Qatar-World PC graph is normal, reasonable... it poses no problems and fits well with what we know from other data.
But this case is completely different: while the legend says normal things, the graph does not correlate neither with the legend nor the expectations.
Regarding the statement in the supplement:
ReplyDelete"the actual plot shows that PC1 separates East Asians from the rest and PC2 Indians (GIH) from the rest, with Europeans and African clumping together in the same corner. "
I believe they meant to say:
"the actual PC1 plot shows that the eigenvector 1 dimension separates East Asians from the rest and the eigenvector 2 dimension separates Indians (GIH) from the rest, with Europeans and African clumping together in the same corner."
"You mean sub-Saharan (as ultra=beyond)?"
ReplyDeleteI mean ultra-Saharan as "beyond the Sahara", yes. I do not mean sub-Saharan as "under the Sahara" because no people lives there (and it's a racist term IMO).
Other possible terms are trans-Saharan (like in "Transalpine Gaul" but can be confused with "trans-Saharan routes", meaning "across" instead of "beyond") and one I use often: "Tropical Africa" (but technically excludes the southernmost tip of Africa, which is subtropical in fact).
I do not mean sub-Saharan as "under the Sahara" because no people lives there (and it's a racist term IMO).
ReplyDeleteIf you mean by the common and academically well established term "sub-Saharan Africa", like me and all other people I know, regions of Africa that lie further south than Sahara, of course people live there. And it isn't remotely racist (it is purely based on cartography, not anything else), also academics use it frequently. And lastly, unlike your alternative, it is direction-neutral.
I wouldn't like to hijack the thread on this matter so you may want to continue this branch of the discussion at Leherensuge: 'Super-Saharan Africa' article, which is a short article I wrote on the matter in 2008. I'm sure that Dienekes and other readers will thank that we divert the branch debate to somewhere else.
ReplyDeleteIn any case: "sub" means "under" or "inferior" and is generally used in a negative sense, like subhuman, subnormal, submissive, subordinate, etc. It is not an "academic" term but one originated in the European mass media in relation to immigration when using the word Black (the traditional term for good or bad) was perceived by some to be "racially charged", so some "genius" journalist some day got a map and, voilá!, invented a new word out of the convention of placing the south at the bottom of maps. Somehow (European media sloppiness as far as I can recall) it became mainstream in the late 80s or rather early 90s but not without some raising our eyebrows and our protests.
Etymologically and geographically it's an incorrect term and IMO has a clear attitude of disrespect. I'd rather use Black Africa, sincerely.
Maju.
ReplyDelete"Etymologically and geographically it's an incorrect term and IMO has a clear attitude of disrespect."
subarctic
subterranean
subsiduary
subconscious
subduction
Are these also terms that must be excised from the English Language.
Africans themselves use the term as in Sub-Saharan Publishers.
There can be only one up and one down and for a while, North has been up. It's a little late to change it.
I don't hear the Australians complaining about being from the "land down under." Or is that also a disrespectful term?
Seriously, the Africans have bigger fish to fry.
Marnie: I have already said that I don't want to continue the discussion on the term "sub-saharan" here because it's not the main subject of discussion and I'm sure that Dienekes is going to get pissed off.
ReplyDeleteSuffice to say that I don't use it for the reasons explained. Anyhow I strongly suspect it's a deformation of "Sud-Saharien/-ano", a valid Romance term (like Sudamérica).
Maju, as you don't want to further this semantic debate, I will finish it by saying that I don't share your super, hyper, over or ultra (I don't know which one is the least racist for you) sensitivity.
ReplyDelete