Nature 441, 1103-1108 (29 June 2006)
Genetic evidence for complex speciation of humans and chimpanzees
Nick Patterson et al.
Abstract
The genetic divergence time between two species varies substantially across the genome, conveying important information about the timing and process of speciation. Here we develop a framework for studying this variation and apply it to about 20 million base pairs of aligned sequence from humans, chimpanzees, gorillas and more distantly related primates. Human–chimpanzee genetic divergence varies from less than 84% to more than 147% of the average, a range of more than 4 million years. Our analysis also shows that human–chimpanzee speciation occurred less than 6.3 million years ago and probably more recently, conflicting with some interpretations of ancient fossils. Most strikingly, chromosome X shows an extremely young genetic divergence time, close to the genome minimum along nearly its entire length. These unexpected features would be explained if the human and chimpanzee lineages initially diverged, then later exchanged genes before separating permanently.
Link
June 30, 2006
Comment on mtDNA from European Neolithic farmers
Albert J. Ammerman, Ron Pinhasi, and Eszter Bánffy have published a comment (Science 30 June 2006:Vol. 312. no. 5782, p. 1875) on the article about the mtDNA of early central European farmers.
On the basis of analysis of ancient DNA from early European farmers, Haak et al. (Reports, 11 November 2005, p. 1016) argued for the Paleolithic ancestry of modern Europeans. We stress that the study is more limited in scope than the authors claim, in part because not all of the skeletal samples date to the time of the Neolithic transition in a given area of Europe.There is also a reply (Science 30 June 2006: Vol. 312. no. 5782, p. 1875) in the same issue.
...Haak et al. (1) report findings on mitochondrial DNA that are of interest, and such work needs to be encouraged. However, the argument they set forth with respect to the ancestry of modern Europeans rests on two working assumptions: (i) that the occurrence of the N1a type among the first farmwomen of Central Europe has its source in the first farmwomen of southeastern Europe (who spread to Central Europe as part of the Neolithic transition) and (ii) that N1a is not present in the local Mesolithic (late hunter-gatherer) populations of Central Europe. Empirical studies have yet to be carried out in either case. More work remains to be done before definite conclusions can be drawn about European ancestry as a whole.
June 29, 2006
Reproductive success of left-handers
Evolution and Human Behavior (Article in Press)
Handedness and reproductive success in two large cohorts of French adults
Charlotte Faurie et al.
Abstract
Left- and right-handers in humans coexist at least since the Paleolithic, and this variation in hand preference has a heritable basis. Because there is extensive evidence of an association between left-handedness and several fitness costs, the persistence of the polymorphism requires an explanation. It is not known whether the frequency of left-handedness in Western societies is stable or not. If the polymorphism is at equilibrium and maintained by frequency dependence, it implies that the fitness of left-handers equals that of right-handers. On the contrary, if left- and right-handers have a different fitness, the polymorphism will evolve. Using two large cohorts of French adults (men and women), we investigated the relations between handedness and several estimators of the reproductive value: marital status, number of sexual partners (of the opposite sex), number of children, and number of grandchildren. Left-handers seem to have disadvantages for some life-history traits, such as marital status (for women) and number of children. For other traits, we observed sex-dependent interactions with socioeconomic status: for high-income categories, left-handed women report less sex partners and left-handed men have more grandchildren. These kinds of interactions are to be expected under the hypothesis that the polymorphism of handedness is stable.
Link
Handedness and reproductive success in two large cohorts of French adults
Charlotte Faurie et al.
Abstract
Left- and right-handers in humans coexist at least since the Paleolithic, and this variation in hand preference has a heritable basis. Because there is extensive evidence of an association between left-handedness and several fitness costs, the persistence of the polymorphism requires an explanation. It is not known whether the frequency of left-handedness in Western societies is stable or not. If the polymorphism is at equilibrium and maintained by frequency dependence, it implies that the fitness of left-handers equals that of right-handers. On the contrary, if left- and right-handers have a different fitness, the polymorphism will evolve. Using two large cohorts of French adults (men and women), we investigated the relations between handedness and several estimators of the reproductive value: marital status, number of sexual partners (of the opposite sex), number of children, and number of grandchildren. Left-handers seem to have disadvantages for some life-history traits, such as marital status (for women) and number of children. For other traits, we observed sex-dependent interactions with socioeconomic status: for high-income categories, left-handed women report less sex partners and left-handed men have more grandchildren. These kinds of interactions are to be expected under the hypothesis that the polymorphism of handedness is stable.
Link
June 28, 2006
The shape of porn actresses
I wonder how they collected the anthropometric data...
Arch Sex Behav. 2006 Jun 24; [Epub ahead of print]
Success Is All in the Measures: Androgenousness, Curvaceousness, and Starring Frequencies in Adult Media Actresses.
Voracek M, Fisher ML.
The debate of whether body-mass index (BMI) or waist-to-hip ratio (WHR) is the primary visual cue to female physical attractiveness (FPA) has generated considerable interest. However, experiments addressing this question typically have limited ecological validity and do not capture the ultimate goal of FPA, which is to elicit male sexual arousal. Hence, using an unobtrusive measures design, we retrieved movie and magazine starring frequencies of 125 adult media actresses from a company's database, operationalized starring frequencies as FPA measures, and tested their relationship to actresses' anthropometric data. Low BMI was related to frequent movie starring, while WHR, waist-to-bust ratio (WBR), and bust size were not. Conversely, low WHR, low WBR, and larger bust size were related to frequent magazine starring, while BMI was not. Visual cues to FPA might be domain-specific, with androgenousness cues salient for attractiveness evaluation of moving bodies and curvaceousness cues salient for posing bodies.
Link
Arch Sex Behav. 2006 Jun 24; [Epub ahead of print]
Success Is All in the Measures: Androgenousness, Curvaceousness, and Starring Frequencies in Adult Media Actresses.
Voracek M, Fisher ML.
The debate of whether body-mass index (BMI) or waist-to-hip ratio (WHR) is the primary visual cue to female physical attractiveness (FPA) has generated considerable interest. However, experiments addressing this question typically have limited ecological validity and do not capture the ultimate goal of FPA, which is to elicit male sexual arousal. Hence, using an unobtrusive measures design, we retrieved movie and magazine starring frequencies of 125 adult media actresses from a company's database, operationalized starring frequencies as FPA measures, and tested their relationship to actresses' anthropometric data. Low BMI was related to frequent movie starring, while WHR, waist-to-bust ratio (WBR), and bust size were not. Conversely, low WHR, low WBR, and larger bust size were related to frequent magazine starring, while BMI was not. Visual cues to FPA might be domain-specific, with androgenousness cues salient for attractiveness evaluation of moving bodies and curvaceousness cues salient for posing bodies.
Link
mtDNA of Mon-Khmer and Mundari
Am J Hum Biol. 2006 Jul-Aug;18(4):461-9.
Asian and non-Asian origins of Mon-Khmer- and Mundari-speaking Austro-Asiatic populations of India.
Kumar V, Langsiteh BT, Biswas S, Babu JP, Rao TN, Thangaraj K, Reddy AG, Singh L, Reddy BM.
In the present study, we analyzed 1,686 samples from 31 tribal populations of India for the mitochondrial DNA 9-base-pair deletion/insertion polymorphism, and characterized them based on the relevant mitochondrial DNA coding-region single nucleotide polymorphisms and hypervariable region I motifs, to test the genetic origins of the ethnically and linguistically heterogeneous Austro-Asiatic tribes of India. A comparative analysis of our results with the existing data suggests multiple origins of Austro-Asiatic tribes in India, and particularly the Asian and non-Asian origins of the Mon-Khmer and the Mundari populations. We also identified a novel subclade of haplogroup B in the Mon-Khmer Khasi tribes that distinguishes them from the Nicobarese, indicating two different waves of migration of the Mon-Khmer tribes in India.
Link
Asian and non-Asian origins of Mon-Khmer- and Mundari-speaking Austro-Asiatic populations of India.
Kumar V, Langsiteh BT, Biswas S, Babu JP, Rao TN, Thangaraj K, Reddy AG, Singh L, Reddy BM.
In the present study, we analyzed 1,686 samples from 31 tribal populations of India for the mitochondrial DNA 9-base-pair deletion/insertion polymorphism, and characterized them based on the relevant mitochondrial DNA coding-region single nucleotide polymorphisms and hypervariable region I motifs, to test the genetic origins of the ethnically and linguistically heterogeneous Austro-Asiatic tribes of India. A comparative analysis of our results with the existing data suggests multiple origins of Austro-Asiatic tribes in India, and particularly the Asian and non-Asian origins of the Mon-Khmer and the Mundari populations. We also identified a novel subclade of haplogroup B in the Mon-Khmer Khasi tribes that distinguishes them from the Nicobarese, indicating two different waves of migration of the Mon-Khmer tribes in India.
Link
June 26, 2006
The Tree of Life is not rooted in the Archaea
A new article rejects the hypothesis that the tree of life on the planet is rooted in the Archaea.
Molecular Biology and Evolution (Advance Access)
Evidence that the Root of the Tree of Life is not within the Archaea
Ryan G. Skophammer et al.
Abstract
The Archaea occupy uncommon and extreme habitats around the world. They manufacture unusual compounds, utilize novel metabolic pathways, and contain many unique genes. Many suspect, due to their novel properties, that the root of the tree of life may be within the Archaea, although there is little direct evidence for this root. Here, using gene insertions and deletions found within protein synthesis factors present in all prokaryotes and eukaryotes, we present statistically significant evidence that the root of life is outside the Archaea.
Link
Molecular Biology and Evolution (Advance Access)
Evidence that the Root of the Tree of Life is not within the Archaea
Ryan G. Skophammer et al.
Abstract
The Archaea occupy uncommon and extreme habitats around the world. They manufacture unusual compounds, utilize novel metabolic pathways, and contain many unique genes. Many suspect, due to their novel properties, that the root of the tree of life may be within the Archaea, although there is little direct evidence for this root. Here, using gene insertions and deletions found within protein synthesis factors present in all prokaryotes and eukaryotes, we present statistically significant evidence that the root of life is outside the Archaea.
Link
June 24, 2006
Expansion of pigs prior to domestication
Proc Biol Sci. 2006 Jul 22;273(1595):1803-10.
Mitochondrial diversity in European and Chinese pigs is consistent with population expansions that occurred prior to domestication.
Fang M, Andersson L.
Mitochondrial DNA (mtDNA) diversity in European and Asian pigs was assessed using 1536 samples representing 45 European and 21 Chinese breeds. Diagnostic nucleotide differences in the cytochrome b (Cytb) gene between the European and Asian mtDNA variants were determined by pyrosequencing as a rapid screening method. Subsequently, 637bp of the hypervariable control region was sequenced to further characterize mtDNA diversity. All sequences belonged to the D1 and D2 clusters of pig mtDNA originating from ancestral wild boar populations in Europe and Asia, respectively. The average frequency of Asian mtDNA haplotypes was 29% across European breeds, but varied from 0 to 100% within individual breeds. A neighbour-joining (NJ) tree of control region sequences showed that European and Asian haplotypes form distinct clusters consistent with the independent domestication of pigs in Asia and Europe. The Asian haplotypes found in the European pigs were identical or closely related to those found in domestic pigs from Southeast China. The star-like pattern detected by network analysis for both the European and Asian haplotypes was consistent with a previous demographic expansion. Mismatch analysis supported this notion and suggested that the expansion was initiated before domestication.
Link
Mitochondrial diversity in European and Chinese pigs is consistent with population expansions that occurred prior to domestication.
Fang M, Andersson L.
Mitochondrial DNA (mtDNA) diversity in European and Asian pigs was assessed using 1536 samples representing 45 European and 21 Chinese breeds. Diagnostic nucleotide differences in the cytochrome b (Cytb) gene between the European and Asian mtDNA variants were determined by pyrosequencing as a rapid screening method. Subsequently, 637bp of the hypervariable control region was sequenced to further characterize mtDNA diversity. All sequences belonged to the D1 and D2 clusters of pig mtDNA originating from ancestral wild boar populations in Europe and Asia, respectively. The average frequency of Asian mtDNA haplotypes was 29% across European breeds, but varied from 0 to 100% within individual breeds. A neighbour-joining (NJ) tree of control region sequences showed that European and Asian haplotypes form distinct clusters consistent with the independent domestication of pigs in Asia and Europe. The Asian haplotypes found in the European pigs were identical or closely related to those found in domestic pigs from Southeast China. The star-like pattern detected by network analysis for both the European and Asian haplotypes was consistent with a previous demographic expansion. Mismatch analysis supported this notion and suggested that the expansion was initiated before domestication.
Link
June 23, 2006
Gene-culture co-evolution of costly punishment
Science 23 June 2006:
Vol. 312. no. 5781, pp. 1767 - 1770
Costly Punishment Across Human Societies
Joseph Henrich et al.
Recent behavioral experiments aimed at understanding the evolutionary foundations of human cooperation have suggested that a willingness to engage in costly punishment, even in one-shot situations, may be part of human psychology and a key element in understanding our sociality. However, because most experiments have been confined to students in industrialized societies, generalizations of these insights to the species have necessarily been tentative. Here, experimental results from 15 diverse populations show that (i) all populations demonstrate some willingness to administer costly punishment as unequal behavior increases, (ii) the magnitude of this punishment varies substantially across populations, and (iii) costly punishment positively covaries with altruistic behavior across populations. These findings are consistent with models of the gene-culture coevolution of human altruism and further sharpen what any theory of human cooperation needs to explain.
Link
Vol. 312. no. 5781, pp. 1767 - 1770
Costly Punishment Across Human Societies
Joseph Henrich et al.
Recent behavioral experiments aimed at understanding the evolutionary foundations of human cooperation have suggested that a willingness to engage in costly punishment, even in one-shot situations, may be part of human psychology and a key element in understanding our sociality. However, because most experiments have been confined to students in industrialized societies, generalizations of these insights to the species have necessarily been tentative. Here, experimental results from 15 diverse populations show that (i) all populations demonstrate some willingness to administer costly punishment as unequal behavior increases, (ii) the magnitude of this punishment varies substantially across populations, and (iii) costly punishment positively covaries with altruistic behavior across populations. These findings are consistent with models of the gene-culture coevolution of human altruism and further sharpen what any theory of human cooperation needs to explain.
Link
June 20, 2006
mtDNA and sheep domestication
Molecular Biology and Evolution (advance access)
Sheep Mitochondrial DNA Variation in European, Caucasian and Central Asian Areas
Miika Tapio et al.
Abstract
Three distinct mitochondrial (mt) maternal lineages (haplotype groups A, B and C) have been found in the domestic sheep. Group B has been observed primarily in European domestic sheep. The European mouflon carries this haplotype group. This could suggest that European mouflon was independently domesticated in Europe, although archaeological evidence supports sheep domestication in the central part of the Fertile Crescent. To investigate this question, we sequenced a highly variable segment of mtDNA in 406 unrelated animals from 48 breeds or local varieties. They originated from a wide area spanning northern Europe and the Balkans to the Altay Mountains in south Siberia. The sample included a representative cross-section of sheep breeds from areas close to the postulated Near Eastern domestication centre and breeds from more distant northern areas. Four (A, B, C and D) highly diverged sheep lineages were observed in Caucasus, three (A, B and C) in Central Asia, and two (A and B) in the eastern fringe of Europe, which included the area north and west of the Black Sea and the Ural Mountains. Only one example of Group D was detected. The other haplotype groups demonstrated signs of population expansion. Sequence variation within the lineages implied Group A to have expanded first. This group was the most frequent type only in Caucasian and Central Asian breeds. Expansion of Group C appeared most recently. The expansion of Group B involving Caucasian sheep took place at nearly the same time as the expansion of Group A. Group B expansion for the eastern European area started approximately 3,000 years after the earliest inferred expansion. An independent European domestication of sheep is unlikely. The distribution of Group A variation as well as other results are compatible with the Near East being the domestication site. Group C and D may have been introgressed later into a domestic stock, but larger samples are needed to infer their geographical origin. The results suggest that some mitochondrial lineages arrived in northern Europe from the Near East across Russia.
Link
Sheep Mitochondrial DNA Variation in European, Caucasian and Central Asian Areas
Miika Tapio et al.
Abstract
Three distinct mitochondrial (mt) maternal lineages (haplotype groups A, B and C) have been found in the domestic sheep. Group B has been observed primarily in European domestic sheep. The European mouflon carries this haplotype group. This could suggest that European mouflon was independently domesticated in Europe, although archaeological evidence supports sheep domestication in the central part of the Fertile Crescent. To investigate this question, we sequenced a highly variable segment of mtDNA in 406 unrelated animals from 48 breeds or local varieties. They originated from a wide area spanning northern Europe and the Balkans to the Altay Mountains in south Siberia. The sample included a representative cross-section of sheep breeds from areas close to the postulated Near Eastern domestication centre and breeds from more distant northern areas. Four (A, B, C and D) highly diverged sheep lineages were observed in Caucasus, three (A, B and C) in Central Asia, and two (A and B) in the eastern fringe of Europe, which included the area north and west of the Black Sea and the Ural Mountains. Only one example of Group D was detected. The other haplotype groups demonstrated signs of population expansion. Sequence variation within the lineages implied Group A to have expanded first. This group was the most frequent type only in Caucasian and Central Asian breeds. Expansion of Group C appeared most recently. The expansion of Group B involving Caucasian sheep took place at nearly the same time as the expansion of Group A. Group B expansion for the eastern European area started approximately 3,000 years after the earliest inferred expansion. An independent European domestication of sheep is unlikely. The distribution of Group A variation as well as other results are compatible with the Near East being the domestication site. Group C and D may have been introgressed later into a domestic stock, but larger samples are needed to infer their geographical origin. The results suggest that some mitochondrial lineages arrived in northern Europe from the Near East across Russia.
Link
June 16, 2006
Positive natural selection in humans
A new review in Science of the evidence for positive natural selection in humans.
Science 16 June 2006:
Vol. 312. no. 5780, pp. 1614 - 1620
Positive Natural Selection in the Human Lineage
P. C. Sabeti et al.
Positive natural selection is the force that drives the increase in prevalence of advantageous traits, and it has played a central role in our development as a species. Until recently, the study of natural selection in humans has largely been restricted to comparing individual candidate genes to theoretical expectations. The advent of genome-wide sequence and polymorphism data brings fundamental new tools to the study of natural selection. It is now possible to identify new candidates for selection and to reevaluate previous claims by comparison with empirical distributions of DNA sequence variation across the human genome and among populations. The flood of data and analytical methods, however, raises many new challenges. Here, we review approaches to detect positive natural selection, describe results from recent analyses of genome-wide data, and discuss the prospects and challenges ahead as we expand our understanding of the role of natural selection in shaping the human genome.
Link
Science 16 June 2006:
Vol. 312. no. 5780, pp. 1614 - 1620
Positive Natural Selection in the Human Lineage
P. C. Sabeti et al.
Positive natural selection is the force that drives the increase in prevalence of advantageous traits, and it has played a central role in our development as a species. Until recently, the study of natural selection in humans has largely been restricted to comparing individual candidate genes to theoretical expectations. The advent of genome-wide sequence and polymorphism data brings fundamental new tools to the study of natural selection. It is now possible to identify new candidates for selection and to reevaluate previous claims by comparison with empirical distributions of DNA sequence variation across the human genome and among populations. The flood of data and analytical methods, however, raises many new challenges. Here, we review approaches to detect positive natural selection, describe results from recent analyses of genome-wide data, and discuss the prospects and challenges ahead as we expand our understanding of the role of natural selection in shaping the human genome.
Link
June 14, 2006
Y chromosomes of Iran
This is a potentially very important high-resolution Y chromosome study of Iran, which was previously studied only with low-resolution markers.
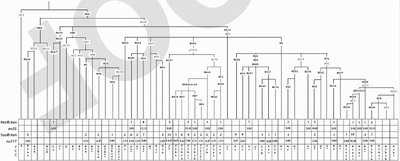
Hum Hered. 2006 Jun 12;61(3):132-143 [Epub ahead of print]
Iran: Tricontinental Nexus for Y-Chromosome Driven Migration.
Regueiro M, Cadenas AM, Gayden T, Underhill PA, Herrera RJ.
Due to its pivotal geographic position, present day Iran likely served as a gateway of reciprocal human movements. However, the extent to which the deserts within the Iranian plateau and the mountain ranges surrounding Persia inhibited gene flow via this corridor remains uncertain. In order to assess the magnitude of this region's role as a nexus for Africa, Asia and Europe in human migrations, high-resolution Y-chromosome analyses were performed on 150 Iranian males. Haplogroup data were subsequently compared to regional populations characterized at similar phylogenetic levels. The Iranians display considerable haplogroup diversity consistent with patterns observed in populations of the Middle East overall, reinforcing the notion of Persia as a venue for human disseminations. Admixture analyses of geographically targeted, regional populations along the latitudinal corridor spanning from Anatolia to the Indus Valley demonstrated contributions to Persia from both the east and west. However, significant differences were uncovered upon stratification of the gene donors, including higher proportions from central east and southeast Turkey as compared to Pakistan. In addition to the modulating effects of geographic obstacles, culturally mediated amalgamations consistent with the diverse spectrum of a variety of historical empires may account for the distribution of haplogroups and lineages observed. Our study of high-resolution Y-chromosome genotyping allowed for an in-depth analysis unattained in previous studies of the area, revealing important migratory and demographic events that shaped the contemporary genetic landscape.
Link

Hum Hered. 2006 Jun 12;61(3):132-143 [Epub ahead of print]
Iran: Tricontinental Nexus for Y-Chromosome Driven Migration.
Regueiro M, Cadenas AM, Gayden T, Underhill PA, Herrera RJ.
Due to its pivotal geographic position, present day Iran likely served as a gateway of reciprocal human movements. However, the extent to which the deserts within the Iranian plateau and the mountain ranges surrounding Persia inhibited gene flow via this corridor remains uncertain. In order to assess the magnitude of this region's role as a nexus for Africa, Asia and Europe in human migrations, high-resolution Y-chromosome analyses were performed on 150 Iranian males. Haplogroup data were subsequently compared to regional populations characterized at similar phylogenetic levels. The Iranians display considerable haplogroup diversity consistent with patterns observed in populations of the Middle East overall, reinforcing the notion of Persia as a venue for human disseminations. Admixture analyses of geographically targeted, regional populations along the latitudinal corridor spanning from Anatolia to the Indus Valley demonstrated contributions to Persia from both the east and west. However, significant differences were uncovered upon stratification of the gene donors, including higher proportions from central east and southeast Turkey as compared to Pakistan. In addition to the modulating effects of geographic obstacles, culturally mediated amalgamations consistent with the diverse spectrum of a variety of historical empires may account for the distribution of haplogroups and lineages observed. Our study of high-resolution Y-chromosome genotyping allowed for an in-depth analysis unattained in previous studies of the area, revealing important migratory and demographic events that shaped the contemporary genetic landscape.
Link
Non-hispanic blacks have the best hearing
Non-Hispanic blacks have best hearing in US, new study shows
Non-Hispanic black adults in the U.S. have on average the best hearing in the nation, a new study shows, with women hearing better than men in general. Overall, the nation's hearing health remains about the same as it was 35 years ago, despite massive changes in society and technology. The results were presented last week at the Acoustical Society of America's spring meeting in Providence, Rhode Island.
William Murphy, Christa Themann, and Mark Stephenson at the National Institute for Occupational Safety and Health (NIOSH) in Cincinnati studied the hearing test results of more than 5,000 U.S. adults aged 20-69 who identified themselves as members of one of three major ethnic groups in the U.S. They studied the adults' "hearing thresholds," the softest sound an individual could hear, over a range of frequencies. They found that non-Hispanic blacks have on average the best hearing thresholds, non-Hispanic whites the worst, with Mexican Americans in between. Women in general had better hearing compared to men.
Comparing the new hearing data to a similar study 35 years ago with adults aged 25-74, the researchers found the median hearing levels in U.S. adults have not changed much; the hearing of U.S. residents is on average not any worse, nor any better than in the early 1970s. This is somewhat surprising because of the greater number of noise sources now present in our society. One potential factor is that hearing protection was not widely available in the early 1970s. Another speculation for the results is that fewer U.S. residents are working in noisy factory jobs, potentially offsetting the effects of newer noise sources. In addition, it is worth noting that the effects of playing portable music players such as now-ubiqitous iPod too loudly might not yet fully be accounted for, since the analyzed data span the years 1999-2004.
The U.S. adults had their hearing tested as part of a more comprehensive study called the National Health and Nutrition Examination Survey (NHANES). In this study, individuals fill out a survey and receive a comprehensive set of tests in a mobile examination center that travels around the U.S.
Hearing loss can be caused by a myriad of factors, such as age, noise exposure (occupational or recreational), developmental syndromes, infectious disease, physical trauma, ototoxic drugs and chemicals, all of which may be influenced by genetic susceptibility. However, it is estimated that at least one third of the cases of hearing impairment stem from overexposure to noise. Estimates of noise exposure in the United States vary, but range from 5 to 30 million persons exposed in the workplace and 16 to 66 million exposed recreationally. Effective prevention programs could therefore make a large impact in reducing then prevalence of hearing loss in the United States.
June 13, 2006
Mellars on human dispersals
Anatomically modern humans appear in the anthropological record long before they spread around the world. Why did it take them so long? I have often repeated in this blog the idea that most humans are descended from a subset of moderns (whom I call the "Afrasians") originating in an eastern African cradle. This small group of Afrasians migrated out of Africa long after the emergence of modern anatomy, and it also spread across Africa where it met other established populations of humans ("Paleoafricans").
A new paper by Paul Mellars in PNAS seems to support the main elements of this thesis. From the paper:
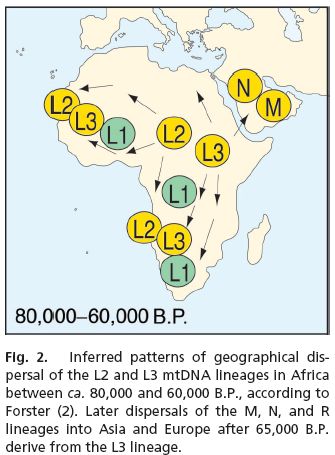
Proc. Natl. Acad. Sci. USA, 10.1073/pnas.0510792103
Why did modern human populations disperse from Africa ca. 60,000 years ago? A new model
Paul Mellars
Recent research has provided increasing support for the origins of anatomically and genetically "modern" human populations in Africa between 150,000 and 200,000 years ago, followed by a major dispersal of these populations to both Asia and Europe sometime after ca. 65,000 before present (B.P.). However, the central question of why it took these populations {approx}100,000 years to disperse from Africa to other regions of the world has never been clearly resolved. It is suggested here that the answer may lie partly in the results of recent DNA studies of present-day African populations, combined with a spate of new archaeological discoveries in Africa. Studies of both the mitochondrial DNA (mtDNA) mismatch patterns in modern African populations and related mtDNA lineage-analysis patterns point to a major demographic expansion centered broadly within the time range from 80,000 to 60,000 B.P., probably deriving from a small geographical region of Africa. Recent archaeological discoveries in southern and eastern Africa suggest that, at approximately the same time, there was a major increase in the complexity of the technological, economic, social, and cognitive behavior of certain African groups, which could have led to a major demographic expansion of these groups in competition with other, adjacent groups. It is suggested that this complex of behavioral changes (possibly triggered by the rapid environmental changes around the transition from oxygen isotope stage 5 to stage 4) could have led not only to the expansion of the L2 and L3 mitochondrial lineages over the whole of Africa but also to the ensuing dispersal of these modern populations over most regions of Asia, Australasia, and Europe, and their replacement (with or without interbreeding) of the preceding "archaic" populations in these regions.
Link
A new paper by Paul Mellars in PNAS seems to support the main elements of this thesis. From the paper:

Proc. Natl. Acad. Sci. USA, 10.1073/pnas.0510792103
Why did modern human populations disperse from Africa ca. 60,000 years ago? A new model
Paul Mellars
Recent research has provided increasing support for the origins of anatomically and genetically "modern" human populations in Africa between 150,000 and 200,000 years ago, followed by a major dispersal of these populations to both Asia and Europe sometime after ca. 65,000 before present (B.P.). However, the central question of why it took these populations {approx}100,000 years to disperse from Africa to other regions of the world has never been clearly resolved. It is suggested here that the answer may lie partly in the results of recent DNA studies of present-day African populations, combined with a spate of new archaeological discoveries in Africa. Studies of both the mitochondrial DNA (mtDNA) mismatch patterns in modern African populations and related mtDNA lineage-analysis patterns point to a major demographic expansion centered broadly within the time range from 80,000 to 60,000 B.P., probably deriving from a small geographical region of Africa. Recent archaeological discoveries in southern and eastern Africa suggest that, at approximately the same time, there was a major increase in the complexity of the technological, economic, social, and cognitive behavior of certain African groups, which could have led to a major demographic expansion of these groups in competition with other, adjacent groups. It is suggested that this complex of behavioral changes (possibly triggered by the rapid environmental changes around the transition from oxygen isotope stage 5 to stage 4) could have led not only to the expansion of the L2 and L3 mitochondrial lineages over the whole of Africa but also to the ensuing dispersal of these modern populations over most regions of Asia, Australasia, and Europe, and their replacement (with or without interbreeding) of the preceding "archaic" populations in these regions.
Link
June 12, 2006
June 10, 2006
Even skin tone is best for women
Women’s skin tone influences perception of beauty, health, age, sociobiologists find
Skin discoloration, uneven tone can add years to perceived age
Philadelphia, Penn. (June 9, 2006) -- Using a revolutionary imaging process, a new study is revealing that wrinkles aren't the only cue the human eye looks for to evaluate age. Scientists at the Ludwig-Boltzmann-Institute for Urban Ethology (Austria) and the Department for Sociobiology/Anthropology at the University of Goettingen (Germany), have shown that facial skin color distribution, or tone, can add, or subtract, as much as 20 years to a woman's age. The study is to be presented at the Human Behavior and Evolution Society (HBES) annual meeting, June 7-11, 2006, in Philadelphia, PA. The study used 3-D imaging and morphing software technologies to remove wrinkles and bone structure from the equation to determine the true impact of facial skin color distribution on the perception of a woman's age, health and attractiveness and is currently in the edit acceptance process with the journal Evolution and Human Behavior.
"Until now, skin's overall homogeneity and color saturation received little attention among behavioral scientists. This study helps us better understand that wrinkles are not the only age cue. Skin tone and luminosity may be a major signal for mate selection and attractiveness, as well as perceived age," says lead researcher Dr. Karl Grammer, Founder and Scientific Director of the Ludwig-Boltzmann-Institute for Urban Ethology, University of Vienna, Austria.
Taking digital photos of 169 Caucasian women aged 10-70, the researchers used specialized morphing software to "drape" each subject's facial skin over a standardized bone structure. Other potential age-defining features such as facial furrows, lines and wrinkles were removed. The subjects who were judged to have the most even skin tone also received significantly higher ratings for attractiveness and health, and were judged to be younger in age.
Tone variances can be caused by several factors including cumulative UV damage (freckles, moles, age spots) natural aging (yellowing, dullness) and skin vascularization (redness). Not surprisingly, the study hinted at a positive correlation between the amount of accumulated photodamage and the amount of uneven skin tone.
"Whether a woman is 17 or 70, the contrast of skin tone plays a significant role in the way her age, beauty and health is perceived," says study co-author Dr. Bernhard Fink, Senior Scientist in the Department for Sociobiology/Anthropology at the University of Goettingen, Germany. "Skin tone homogeneity can give visual clues about a person's health and reproductive capability, so an even skin tone is considered most desirable. In this study, we found cumulative UV damage influences skin tone dramatically, giving women yet another reason to prevent UV-related skin damage or try to correct past damage that is causing uneven skin tone."
Skin discoloration, uneven tone can add years to perceived age
Philadelphia, Penn. (June 9, 2006) -- Using a revolutionary imaging process, a new study is revealing that wrinkles aren't the only cue the human eye looks for to evaluate age. Scientists at the Ludwig-Boltzmann-Institute for Urban Ethology (Austria) and the Department for Sociobiology/Anthropology at the University of Goettingen (Germany), have shown that facial skin color distribution, or tone, can add, or subtract, as much as 20 years to a woman's age. The study is to be presented at the Human Behavior and Evolution Society (HBES) annual meeting, June 7-11, 2006, in Philadelphia, PA. The study used 3-D imaging and morphing software technologies to remove wrinkles and bone structure from the equation to determine the true impact of facial skin color distribution on the perception of a woman's age, health and attractiveness and is currently in the edit acceptance process with the journal Evolution and Human Behavior.
"Until now, skin's overall homogeneity and color saturation received little attention among behavioral scientists. This study helps us better understand that wrinkles are not the only age cue. Skin tone and luminosity may be a major signal for mate selection and attractiveness, as well as perceived age," says lead researcher Dr. Karl Grammer, Founder and Scientific Director of the Ludwig-Boltzmann-Institute for Urban Ethology, University of Vienna, Austria.
Taking digital photos of 169 Caucasian women aged 10-70, the researchers used specialized morphing software to "drape" each subject's facial skin over a standardized bone structure. Other potential age-defining features such as facial furrows, lines and wrinkles were removed. The subjects who were judged to have the most even skin tone also received significantly higher ratings for attractiveness and health, and were judged to be younger in age.
Tone variances can be caused by several factors including cumulative UV damage (freckles, moles, age spots) natural aging (yellowing, dullness) and skin vascularization (redness). Not surprisingly, the study hinted at a positive correlation between the amount of accumulated photodamage and the amount of uneven skin tone.
"Whether a woman is 17 or 70, the contrast of skin tone plays a significant role in the way her age, beauty and health is perceived," says study co-author Dr. Bernhard Fink, Senior Scientist in the Department for Sociobiology/Anthropology at the University of Goettingen, Germany. "Skin tone homogeneity can give visual clues about a person's health and reproductive capability, so an even skin tone is considered most desirable. In this study, we found cumulative UV damage influences skin tone dramatically, giving women yet another reason to prevent UV-related skin damage or try to correct past damage that is causing uneven skin tone."
June 08, 2006
Why men smile less than women
Physiol Behav. 2006 Jun 2; [Epub ahead of print]
Gender differences in smiling: An evolutionary neuroandrogenic theory.
Ellis L.
Studies have found that, under a wide variety of social circumstances, females are more likely than males to smile. The present article offers a theoretical explanation for this difference based on the premise that testosterone (along with other sex hormones) has evolved the tendency to alter brain functioning in ways that inhibit male smiling, especially during their most reproductively active years. Underlying the theory are the assumptions that (a) females have been naturally selected for preferring to mate with males who have the ability to assist in long-term child rearing primarily by provisioning resources, that (b) males partially accommodate this female preference by competing with rival males who are also vying for resources with which to attract mates, and that (c) male smiling interferes with their ability to most effectively intimidate rivals. If this reasoning is correct, genes must be involved in promoting the tendency to compete for resources, the most likely location for which would be on the Y-chromosome. According to the present theory, these genes operate in part by inhibiting social signals of fear and submissiveness. An additional element of the theory asserts that testosterone alters brain functioning in ways that shift the neocortex away from the left (more "prosocial and friendly") hemisphere toward the right (less "prosocial and friendly") hemisphere. Current evidence bearing on the theory is reviewed and a number of largely untested hypotheses are derived from the theory for future assessment of its predictive power.
Link
Gender differences in smiling: An evolutionary neuroandrogenic theory.
Ellis L.
Studies have found that, under a wide variety of social circumstances, females are more likely than males to smile. The present article offers a theoretical explanation for this difference based on the premise that testosterone (along with other sex hormones) has evolved the tendency to alter brain functioning in ways that inhibit male smiling, especially during their most reproductively active years. Underlying the theory are the assumptions that (a) females have been naturally selected for preferring to mate with males who have the ability to assist in long-term child rearing primarily by provisioning resources, that (b) males partially accommodate this female preference by competing with rival males who are also vying for resources with which to attract mates, and that (c) male smiling interferes with their ability to most effectively intimidate rivals. If this reasoning is correct, genes must be involved in promoting the tendency to compete for resources, the most likely location for which would be on the Y-chromosome. According to the present theory, these genes operate in part by inhibiting social signals of fear and submissiveness. An additional element of the theory asserts that testosterone alters brain functioning in ways that shift the neocortex away from the left (more "prosocial and friendly") hemisphere toward the right (less "prosocial and friendly") hemisphere. Current evidence bearing on the theory is reviewed and a number of largely untested hypotheses are derived from the theory for future assessment of its predictive power.
Link
June 06, 2006
Y chromosomes in northern island Melanesia
Molecular Biology and Evolution (advance access)
Unexpected NRY chromosome variation in Northern Island Melanesia
Laura Scheinfeldt et al.
Abstract
To investigate the paternal population history of populations in Northern Island Melanesia, 685 paternally unrelated males from 36 populations in this region and New Guinea were analyzed at 14 regionally informative binary markers and seven short-tandem-repeat loci from the non-recombining portion of the Y chromosome. Three newly defined binary markers (K6-P79, K7-P117, and M2-P87) aided in identifying considerable heterozygosity that would have otherwise gone undetected. Judging from their geographic distributions and network analyses of their associated short-tandem-repeat profiles, four lineages appear to have developed in this region and to be of considerable age: K6-P79, K7-P117, M2-P87, and M2a-P22. The origins of K5-M230 and M-M4 are also confirmed as being located further west, probably in New Guinea. In the 25 adequately sampled populations, the number of different haplogroups ranged from two in the single most isolated group (the Aita of Bougainville), to nine, and measures of molecular diversity were generally not particularly low. The resulting pattern contradicts earlier findings that suggested far lower male-mediated diversity and gene exchange rates in the region. However, these earlier studies had not included the newly defined haplogroups. We could only identify a very weak signal of recent male Southeast Asian genetic influence (<10%), which was almost entirely restricted to Austronesian (Oceanic) speaking groups. This contradicts earlier assumptions on the ancestral composition of these groups and requires a revision of hypotheses concerning the settlement of the islands of the central Pacific, which commenced from this region.
Link
Unexpected NRY chromosome variation in Northern Island Melanesia
Laura Scheinfeldt et al.
Abstract
To investigate the paternal population history of populations in Northern Island Melanesia, 685 paternally unrelated males from 36 populations in this region and New Guinea were analyzed at 14 regionally informative binary markers and seven short-tandem-repeat loci from the non-recombining portion of the Y chromosome. Three newly defined binary markers (K6-P79, K7-P117, and M2-P87) aided in identifying considerable heterozygosity that would have otherwise gone undetected. Judging from their geographic distributions and network analyses of their associated short-tandem-repeat profiles, four lineages appear to have developed in this region and to be of considerable age: K6-P79, K7-P117, M2-P87, and M2a-P22. The origins of K5-M230 and M-M4 are also confirmed as being located further west, probably in New Guinea. In the 25 adequately sampled populations, the number of different haplogroups ranged from two in the single most isolated group (the Aita of Bougainville), to nine, and measures of molecular diversity were generally not particularly low. The resulting pattern contradicts earlier findings that suggested far lower male-mediated diversity and gene exchange rates in the region. However, these earlier studies had not included the newly defined haplogroups. We could only identify a very weak signal of recent male Southeast Asian genetic influence (<10%), which was almost entirely restricted to Austronesian (Oceanic) speaking groups. This contradicts earlier assumptions on the ancestral composition of these groups and requires a revision of hypotheses concerning the settlement of the islands of the central Pacific, which commenced from this region.
Link
100,000 year old Neandertal mtDNA
A letter in Current Biology reports on a 100,000 year old mtDNA sequence. An excerpt:
Current Biology Volume 16, Issue 11 , June 2006, Pages R400-R402
Revisiting Neandertal diversity with a 100,000 year old mtDNA sequence
Ludovic Orlando et al.
Link
The pairwise distance distributions within humans, as well as between humans and Neandertals, are closer and overlap more extensively for more recent Neandertals (p < 0.001) than for the Scladina specimen (Figure 2). While the diversity of the more recent Neandertals is similar to that of modern humans worldwide, the sequence from Scladina reveals that more divergent Neandertal haplotypes existed before 42,000 years ago. This could suggest that Neandertals experienced genetic drift as demographic bottlenecks eliminated the phylogenetically more recent (i.e. less expanded) haplotypes from populations. Consequently, the most likely conserved Neandertal haplotypes could also be the phylogenetically most ancient (i.e. the most closely related to the common ancestor of modern humans and Neandertals). This could explain the shift towards modern human pairwise distributions observed between 100,000 and 40,000 years ago. Whatever this shift should be related to cohabitation, climatic changes, or any subdivision of populations, the Scladina sequence has revealed that the genetic diversity of Neandertals has been underestimated. Thus, more Neandertal sequences than the six presently available and longer than 100 bp are needed to fully understand the extent of the past diversity of Neandertals.
Current Biology Volume 16, Issue 11 , June 2006, Pages R400-R402
Revisiting Neandertal diversity with a 100,000 year old mtDNA sequence
Ludovic Orlando et al.
Link
June 03, 2006
Amazonian knowledge of geometry (revisited)
The following letter to Science are in response to this story about the supposed knowledge of geometry exhibited by a tribe of uncivilized Amazonians. As the author correctly points out, the ability to recognize geometrical figures does not constitute knowledge of geometry.
Science 2 June 2006:
Vol. 312. no. 5778, pp. 1309 - 1310
DOI: 10.1126/science.312.5778.1309c
Examining Knowledge of GeometryLink
In their Report "Core knowledge of geometry in an Amazonian indigene group" (20 Jan., p. 381), S. Dehaene et al. present evidence that an isolated Amazonian group, the Mundurukú, are able to understand geometric concepts. They state that geometry constitutes "a core set of intuitions present in all humans." I disagree with the basic concept of this investigation.
The central feature of Euclidean geometry is its demonstrative character and its logical structure, rather than graphical pictures of triangles, circles, etc. This logical system is built upon two pillars: (i) the concept of the "theoretical object," e.g., the abstract metaphysical idea of a circle, rather than a real constructed circle; and (ii) the deductive mathematical proof, based purely on axioms and postulates.
Other civilizations dealt with geometrical figures in a more intuitive way, and their activities cannot be characterized as geometry in the Euclidean sense. Ancient civilizations other than the Greeks did not develop a demonstrative geometry. For example, the ancient Chinese never developed a theoretical geometry (1-3).
The topic being investigated by Dehaene et al. is simply pattern recognition. It is by no means surprising that the people tested recognized different geometric figures, since they can recognize, e.g., human faces and identify different species of tree by their silhouettes.
Karl Wulff
Geschamp 15
D-29640 Schneverdingen, Germany
E-mail: karl.ah.wulff@t-online.de
References
J. Needham, Science and Civilisation in China, vol. 3, Mathematics and the Science of the Heavens and the Earth (Cambridge University Press, Cambridge, 1959), p. 91.
G. F. Leibniz, Novissima Sinica (ed. 2, 1699), section 9.
C. Cullen, Astronomy and Mathematics in Ancient China: The Zhoubi Suanjing (Cambridge University Press, Cambridge, 1996), pp. 77, 219.
Science 2 June 2006:
Vol. 312. no. 5778, pp. 1309 - 1310
DOI: 10.1126/science.312.5778.1309c
June 01, 2006
Archaic contribution to modern human populations
PLoS Genetics has an article on the possible contribution of genetic material from archaic humans into the modern human gene pool. I have often before stressed the contribution of Paleoafrican populations into the more recent branch of the sapiens lineage ("Afrasians") which emerged in eastern Africa. This new study seems to support the idea of absorption of older African populations by modern humans.
It is clear that the small early modern human population must have inhabited a correspondingly small geographical region, so it is not surprising that in their movements within Africa they would have interbred with the pre-existing humans. After all, humans lived in Africa for a long time before the emergence of the moderns, and there is no reason to believe that all the African branches of humanity were wiped out to be replaced by the advancing moderns. I predict that in the coming years, we will learn much more about the different strata of genetic ancestry contained in Africans, as well as Europeans and East Asians.
Note, also, that there is no candidate for the source population of the archaic contribution of West Africans. This, again, is not surprising, because western Africa has a much less advantageous climate than eastern Africa for bone preservation, in addition to being less well researched. Even in Europe, where anthropological science is the oldest, and cave surveys have been numerous, there are still only a handful of well-preserved Neanderthal specimens. Hopefully, some of the archaics of Africa remain to be discovered.
From the paper:
Possible ancestral structure in human populations
Vincent Plagno, Jeff D. Wall
PROVISIONAL ABSTRACT
Determining the evolutionary relationships between fossil hominid groups such as Neanderthals and modern humans has been a question of enduring interest in human evolutionary genetics. Here we present a new method for addressing whether archaic human groups contributed to the modern gene pool (called ancient admixture) using the patterns of variation in contemporary human populations. Our method improves on previous work by explicitly accounting for recent population history before performing the analyses. Using sequence data from the Environmental Genome Project, we find strong evidence for ancient admixture in both a European and a West African population (p ~ 10^{-7}), with contributions to the modern gene pool of at least 5%. While Neanderthals form an obvious archaic source population candidate in Europe, there is not yet a clear source population candidate in West Africa.
Link
It is clear that the small early modern human population must have inhabited a correspondingly small geographical region, so it is not surprising that in their movements within Africa they would have interbred with the pre-existing humans. After all, humans lived in Africa for a long time before the emergence of the moderns, and there is no reason to believe that all the African branches of humanity were wiped out to be replaced by the advancing moderns. I predict that in the coming years, we will learn much more about the different strata of genetic ancestry contained in Africans, as well as Europeans and East Asians.
Note, also, that there is no candidate for the source population of the archaic contribution of West Africans. This, again, is not surprising, because western Africa has a much less advantageous climate than eastern Africa for bone preservation, in addition to being less well researched. Even in Europe, where anthropological science is the oldest, and cave surveys have been numerous, there are still only a handful of well-preserved Neanderthal specimens. Hopefully, some of the archaics of Africa remain to be discovered.
From the paper:
While the putative source population may not be as obvious as in Europe (Neanderthals), the fossil record shows that transitional forms of Homo were widespread in Africa, even after the time of emergence of modern humans. Other genetic studies have also found evidence for ancient structure in African [28, 29, 30]. In two of the three studies [28, 30], the divergent lineage was found only in Pygmies, which suggests that the African population source differs from the European one.PLoS Genetics (early online)
Possible ancestral structure in human populations
Vincent Plagno, Jeff D. Wall
PROVISIONAL ABSTRACT
Determining the evolutionary relationships between fossil hominid groups such as Neanderthals and modern humans has been a question of enduring interest in human evolutionary genetics. Here we present a new method for addressing whether archaic human groups contributed to the modern gene pool (called ancient admixture) using the patterns of variation in contemporary human populations. Our method improves on previous work by explicitly accounting for recent population history before performing the analyses. Using sequence data from the Environmental Genome Project, we find strong evidence for ancient admixture in both a European and a West African population (p ~ 10^{-7}), with contributions to the modern gene pool of at least 5%. While Neanderthals form an obvious archaic source population candidate in Europe, there is not yet a clear source population candidate in West Africa.
Link
