Most interestingly, there is evidence for gene flow between the MA-1 sample and Native Americans, which makes sense as these are Siberians of the period leading up to the initial colonization of the Americas. The interesting thing is that MA-1 does not appear to be East Eurasian, as proven by the test D(Papuan, Han; Sardinian, MA-1) which is non-significant, so MA-1 is not more closely related to Han than to Papuans (which is true for modern native Americans). So, it seems that the gene flow between MA-1 and Native Americans was towards Native Americans from MA-1 and not vice versa.
It is fascinating that such a sample could be found so far east at so early a time. Both Y-chromosome R and mtDNA haplogroup U are very rare east of Lake Baikal which has been considered a limit of west Eurasian influence into east Eurasia. And, indeed, both these haplogroups are absent in Native Americans, so it is not yet clear how Native Americans (who belong to Y-chromosome haplogroups Q and C and mtDNA haplogroups A, B, C, D, X) are related to these Paleolithic Siberians. The obvious candidate for this relationship is Y-chromosome haplogroup P (the ancestor of Q and R). So, perhaps Q-bearing relatives of the R-bearing Mal'ta population settled the Americas.
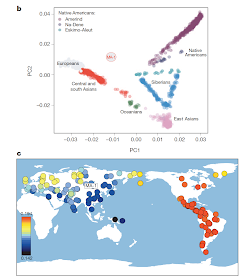
In any case, this is an extremely important sample, as its position in "no man's land" in the PCA plot (left) demonstrates, between Europeans and native Americans but close to no modern population.
Its closest present-day relatives are indicated in (c), with Native Americans (red) being the closest, and a scattering of boreal populations from the Atlantic to the Pacific (but not in the vicinity of Lake Baikal) next in line (yellow).
This distribution clearly related to the evidence for admixture in Europe adduced in two other recent papers, although the question of who went where and when remains to be resolved. Was MA-1 part of an intrusive western population encroaching on east Eurasians? Or did MA-1 lookalikes arrive as first settlers in empty territory, later ceding this space to east Eurasians from, perhaps, China? Did the two mix in Siberia or did they arrive in the Americas in separate migrations and mix there? And, how does this all relate to events in Europe in the far west?
UPDATE: Razib makes an excellent point:
Also, can we now finally bury the debate when east and west Eurasians diverged? Obviously it can’t have been that recent if a >20,000 year old individual had closer affinity to western populations.We already knew that Tianyuan was more Asian than European, so I think west Eurasians diverged from the rest >40 thousand years ago. But, Tianyuan was so early that its precise relationships to different Asian groups could not be determined. So, I'd say it's a good guess that east-west split off before 40 thousand years in Eurasia.
Nature (2013) doi:10.1038/nature12736
Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans
Maanasa Raghavan, Pontus Skoglund et al.
The origins of the First Americans remain contentious. Although Native Americans seem to be genetically most closely related to east Asians1, 2, 3, there is no consensus with regard to which specific Old World populations they are closest to4, 5, 6, 7, 8. Here we sequence the draft genome of an approximately 24,000-year-old individual (MA-1), from Mal’ta in south-central Siberia9, to an average depth of 1×. To our knowledge this is the oldest anatomically modern human genome reported to date. The MA-1 mitochondrial genome belongs to haplogroup U, which has also been found at high frequency among Upper Palaeolithic and Mesolithic European hunter-gatherers10, 11, 12, and the Y chromosome of MA-1 is basal to modern-day western Eurasians and near the root of most Native American lineages5. Similarly, we find autosomal evidence that MA-1 is basal to modern-day western Eurasians and genetically closely related to modern-day Native Americans, with no close affinity to east Asians. This suggests that populations related to contemporary western Eurasians had a more north-easterly distribution 24,000 years ago than commonly thought. Furthermore, we estimate that 14 to 38% of Native American ancestry may originate through gene flow from this ancient population. This is likely to have occurred after the divergence of Native American ancestors from east Asian ancestors, but before the diversification of Native American populations in the New World. Gene flow from the MA-1 lineage into Native American ancestors could explain why several crania from the First Americans have been reported as bearing morphological characteristics that do not resemble those of east Asians2, 13. Sequencing of another south-central Siberian, Afontova Gora-2 dating to approximately 17,000 years ago14, revealed similar autosomal genetic signatures as MA-1, suggesting that the region was continuously occupied by humans throughout the Last Glacial Maximum. Our findings reveal that western Eurasian genetic signatures in modern-day Native Americans derive not only from post-Columbian admixture, as commonly thought, but also from a mixed ancestry of the First Americans.
Link


"Also, can we now finally bury the debate when east and west Eurasians diverged? Obviously it can’t have been that recent if a >20,000 year old individual had closer affinity to western populations.
ReplyDeleteWe already knew that Tianyuan was more Asian than European, so I think west Eurasians diverged from the rest >40 thousand years ago. But, Tianyuan was so early that its precise relationships to different Asian groups could not be determined. So, I'd say it's a good guess that east-west split off before 40 thousand years in Eurasia."
Did you forget about Oceania who might be more closely related to east Asians than east Asians are to west Eurasians. There are multiple over 40,000 year old human remains in Europe. All specifically west Eurasian descendants of mtDNA R so U, RO, and R2'JT. Are estimated to be over 50,000 years old. I would say the splits between non Africans(don't forget about Oceania) happened over 60,000 years ago.
"Or did MA-1 lookalikes arrive as first settlers in empty territory, later ceding this space to east Eurasians from, perhaps, China?"
ReplyDeleteI have been arguing that at the previous post on the subject.
"Did the two mix in Siberia or did they arrive in the Americas in separate migrations and mix there?"
At the same post I have been arguing in favour of mixing in Siberia.
"Similarly, we find autosomal evidence that MA-1 is basal to modern-day western Eurasians and genetically closely related to modern-day Native Americans, with no close affinity to east Asians".
Surely that really does support the idea that Native Americans are an admixed population, in spite of what some still maintain. And this:
"Furthermore, we estimate that 14 to 38% of Native American ancestry may originate through gene flow from this ancient population".
So Native Americans can only be somewhere between 86 and 62% Mongoloid.
"Gene flow from the MA-1 lineage into Native American ancestors could explain why several crania from the First Americans have been reported as bearing morphological characteristics that do not resemble those of east Asians"
Exactly.
Isn't this the OLDEST KNOWN case of y-chromosome R?
ReplyDeleteWhere do the Sami and Finns cluster in this PCA plot? I would think some of the ancestral components in those populations might be related with this North Eurasian H-G population.
ReplyDeleteThe study posits that Native Americans were admixed with this population after their 'divergence from East Eurasians,' but exactly what divergence are they talking about? Native Americans share no principle hg Y with East Eurasians but for a set of clades - C, Q, and R - that are easier to explain via co-admixture with third party populations. Yes, there are shared hgs, but I find the use of the term 'divergence' highly problematic when the two populations are not principally descended from the same ancestors and when no 'parent population' exhibiting the bulk of the shared haplotypes has ever been detected.
ReplyDeleteThe absence of Y hg NO, and for that matter, NO's ancestors in Native Americans has never ceased to perplex me when hearing that East Eurasians and Native Americans 'diverged' tens of thousands of years ago. Setting C on the side, both Q and R belong to the P macro clade, which are on the West Eurasian side of the MNOPS divide. It never made me comfortable that paleo geneticists kept talking about a simple two way split between West and East Eurasians, followed by another split between East Eurasians and Native Americans. It's obvious the story isn't that simple.
As for EDAR, the discussion of which I missed in the October release, two thoughts arise - first, that the variant looks to have had a selective advantage. Second, that in case it did have a selective advantage, then using it to chart the ancient migrations of hgs is not going to be straight forward. In the classic argument where EDAR conferred an adaptive advantage in northerly climates, then the simple fact that it is present in a higher % among Native Americans than East Eurasians does not necessarily indicate that it emerged among Native Americans - only that the environment in which Native Americans dwelt led to higher selection for it, which was then amplified by the founders' effect which produced the Native American genetic landscape.
All in all, this result corroborates several other results that indicate south and central Siberia were not populated by East Eurasians, and specifically NO populations, till rather late - ie in the Neolithic due to expansions from the east. At the same time, recent research by Chinese paleo geneticists have found the mid Neolithic presence of NO - especially N - in what is now Northeast China and eastern Inner Mongolia. I surmise O was to the south of N, though how south I am not yet sure. The picture I thereby glean is one of what later became the Mongolian steppes being the major natural barrier between NO and P.
Such a picture also indicates that NO did not arrive in East Eurasia via Siberia and Mongolia. Indeed, in absence of new evidence, it never crossed north of Inner Mongolia on its way east, and the southward path through South Asia and Tibet is looking increasingly juicy for the arrival of the main ancestor population of East Eurasians. Subsequent expansion took place in and around China before waves of northward pushes spread first N and then O into Siberia, Northeast Asia, and West Eurasia.
The Mongoloid type itself, in this case, looks to be just another areal mishmash of adaptive and decorative genes, and not the touchstone of racial divergence 19th century anthropologists believed that it was.
'The interesting thing is that MA-1 does not appear to be East Eurasian, as proven by the test D(Papuan, Han; Sardinian, MA-1) which is non-significant, so MA-1 is not more closely related to Han than to Papuans (which is true for modern native Americans). So, it seems that the gene flow between MA-1 and Native Americans was towards Native Americans from MA-1 and not vice versa."
ReplyDeleteThis is based on a misunderstanding. Only Eskimo-Aleuts and Na-Dene are closer to Han than they are to Mal'ta (p. 198, Suppl. Mat.) They carry the other New World ancestral component that's different from the First American component . The First American component didn't come from East Asians. And it's precisely that First American component that's found in Mal'ta. Unless Amerindians, save Eskimo-Aleuts and Na-Dene, are 100% West Eurasians (and the paper assigns only a minor West Eurasian ancestry to Amerindians), there's no way they can be a mix of East Asians and West Eurasians.
http://anthropogenesis.kinshipstudies.org/2013/11/ancient-dna-from-malta-and-afontova-gora-a-full-account/
"In any case, this is an extremely important sample, as its position in 'no man's land' in the PCA plot (left) demonstrates, between Europeans and native Americans but close to no modern population."
ReplyDeleteIt looks closer to "South and Central Asians" than Europeans to me. Is there any link to modern Middle Eastern populations?
mtDNA X2 in Native Americans is proof their ancestors mixed with west Eurasians probably while still in Asia or very early in America. Is the west Eurasian ancestry in these Palaeolithic Siberians more related to any modern west Eurasians?
ReplyDeleteI would bet it is somehow connected to North Euro-west Asian in globe13. Which are extremely related to each other. Other Dodoecade tests show similar groups with names like Atlantic Baltic and west Asian which are also extremely related to each other. Obviously some close common ancestry between Europeans and west Asians.
We know North Euro dominated at least some pre Neolithic Europeans because of ancient samples. Today west Asian is over 50% in Caucuses and around 45% from Turkey to Pakistan and decreases as you go south from Iraq. Europe and Iran-Pakistan area are the closest spots to Siberia.
I would think that Mexicans and other Hispanics would be closest to these Palaeolithic Siberians. Because they have mixed Native Americans and Spanish(west Eurasian) ancestry.
Is it that these Palaeolithic Siberians had mixed ancestry from west Eurasians and Native American like people? Or is it that they were a mix of west Eurasian and something? else and their population is partly ancestral to modern Native Americans.
Archeology has long indicated that Siberia was settled from west to east in the 37-35 kya time frame, so this Mal'ta individual was derived (for ~15 ky) away from the-then developing W Eurasian populations. In agreement with this picture, his U lineage is highly derived (and different from extant haplogroups), and his R lineage is somewhat derived (and different from extant haplogroups, but fairly close to the R1/ R2 split). I think that can explain the isolated position on the PC plot (wished they had shown PC3, on a subset of relevant populations, though).
ReplyDeleteI think it is fair to assume that other than making a contribution to Native Americans, this Siberian population largely died out / was replaced by the post-LGM East Asian wedge I have talked about, before (also visible in the heat map).
Based on a couple of papers (that I need to re-read), the authors state as a fact that this individual predates the E Asian - N American split. I find that highly unlikely; clearly, different subregions in NE Asia were settled in the 35ky to 30ky time frame, so the split most likely predates Mal'ta boy by about 5k to 10k years. This is consistent with the idea that the admixture went underway farther East, in extreme E Siberia and Beringia, with a brother group (Q, X) of Mal'ta boy, and not his population, directly.
So, IMO (as I stated before) a proto-Mongoloid admixture for the first Americans, and then successive Mongoloid admixtures for Na Dene and Inuits.
I paid for and read the entire paper and supplement. It supplies a lot of corroborative evidence for Terry T and my arguments in other threads.
ReplyDeleteFirst, there's a strong correlation between the Dodecad k=4 Amerindian component and the relationship of individual European populations to the Mal'ta sample.
There's not much evidence of pan-West Eurasian backflow from Mal'ta, with the Amerindian affinities of all West Eurasians (and Willerslev specifies this also includes Middle-Easterners) being better explained as an expression of relatedness and not actual admixture of Amerindians (or even the West Eurasian progenitors of Amerindians back) into West Eurasia.
NW Siberians and NE Europeans are the most closely related non-Amerindian/extreme NE Siberian populations to the boy. It's not mentioned explicitly, but I previously argued that Mal'ta-like ancestry probably did return to the NE portion of Europe via Finno-Ugric movements (although these genes came mixed with an East Asian component). That seems to be the case, because while those two populations (NE Euros and NW Siberians) are more closely related to the boy, of the standard HGDP Europeans Orcadians are the European population most asymmetrically related to Native Americans vs. East Asians, meaning they're the population best described as being much more related to Native Americans than to East Asians.
It seems reasonable to suggest that mesolithic Europeans originally had a strong relation to the West Eurasian component in Amerindians but not to East Asians. This relation was later bolstered in parts of Europe by extremely close Mal'ta (and therefore Amerindian) relatives from inner Asia moving west into NE Europe, but now carrying haplogroup N and its related genes.
Final observation for now: Treemix shows admixture from Mal'ta to Karitiana to be the second most important admixture event following Denisovan admixture into Papuans.
"This relation was later bolstered in parts of Europe by extremely close Mal'ta (and therefore Amerindian) relatives from inner Asia moving west into NE Europe, but now carrying haplogroup N and its related genes."
ReplyDeleteI doubt that Y DNA N(specifically N1c) was brought to northeast Europe with Mal'ta like people. I have seen in many other autosomal DNA results that northeast Europeans have obviously modern Siberian ancestry not Mal'ta like. Mal'ta also had it seems from K=9 more south Asian than European ancestry.
@John Rudmin
ReplyDelete"Isn't this the OLDEST KNOWN case of y-chromosome R?"
Yes.
I think the oldest known R before this Sibirian was a R1a of a Corded Ware guy in Eastern Germany.
What shows how rare it was (so far) to succesfully estaminate Y-DNA from corpses older than say 2K years while its easy to estaminate mtDNA from 50K year old corpses.
I would think, Y-DNA from a >20K year old corpse would have been declared "absolutely impossible" only 5 years ago.
I'm pleased to see veryone except German accepts the findings at face value. Except for one comment from Eurologist:
ReplyDelete"So, IMO (as I stated before) a proto-Mongoloid admixture for the first Americans"
Proto-Mongoloid? Come off the weed. Surely it is obvious now that the the first Americans were an admixed population. Nothing 'proto- about them except they were proto-Americans. But I agree completely with this:
"the authors state as a fact that this individual predates the E Asian - N American split. I find that highly unlikely"
Lathdrinor covers that aspect well, above.
"The First American component didn't come from East Asians. And it's precisely that First American component that's found in Mal'ta".
Yes, that's what the paper says.
"Unless Amerindians, save Eskimo-Aleuts and Na-Dene, are 100% West Eurasians (and the paper assigns only a minor West Eurasian ancestry to Amerindians), there's no way they can be a mix of East Asians and West Eurasians".
So '14 to 38% of Native American ancestry may originate through gene flow from this ancient population' is minor? I can't for the life of me imagine why you demand that Native Americans cannot be a hybrid because they are not 100% West Eurasian.
@Hamarfox
ReplyDelete"but I previously argued that Mal'ta-like ancestry probably did return to the NE portion of Europe via Finno-Ugric movements"
Or the massive Finno Ugric migrations to Britain? (there was no, but still that guy is related to Brits.
I recall former experiments to calculate the timespan of the admixture event.
those calculations said, Amerindian admixture in northern Europeans is from 17K years ago and exists in all Europeans, in northern Europeans twice as much as in southern ones while there is only a tiny difference between Irish and Russian.
The same calculator said, East Asian admixture in North-East Europeans is from 2500 years ago and exists in Russians but zero in Irish.
"It never made me comfortable that paleo geneticists kept talking about a simple two way split between West and East Eurasians, followed by another split between East Eurasians and Native Americans. It's obvious the story isn't that simple".
ReplyDeleteObvious. I don't know if anyone checked the Razib link, but I really liked this comment there:
"Reticulation. Graphs. Admixture. These words all point to the reality that rather than being the culmination of deep rooted regional populations which date back to the depths of the Pleistocene, most modern humans are recombinations of ancient lineages. On the grandest scale this is illustrated by the evidence of ‘archaic’ ancestry in modern humans. But even more pervasively we see evidence of widespread admixture between distinct lineages which are major world populations which we think of as archetypes".
I am certain the same process goes right back to the emergence of the Homo genus from Australopithecus. The old idea that each new species represents the expansion of a single, 'superior' population is a hangover from Victorian racial and economic ideas: the 'survival of the fittest'. Hopefully the emerging lines of research will lay to rest those old Victorian ideas.
"The Mongoloid type itself, in this case, looks to be just another areal mishmash of adaptive and decorative genes, and not the touchstone of racial divergence 19th century anthropologists believed that it was".
Yes. Every 'race' is the product of admixture. However in the case of the Mongoloid phenotype I am sure the combination of genes came together in a reasonably confined region, and expanded from there, mixing with other populations as it went. And, in turn, becoming admixed with those incoming populations.
This comment has been removed by the author.
ReplyDelete@terryT
ReplyDeleteI know you're trying to pick a fight again. Ok, here you go.
"I'm pleased to see veryone except German accepts the findings at face value."
People follow the opinions of authoritative others. They don't look at the data. I do. And I'm not the only one. The David Reich lab doesn't accept this paper's conclusion that Amerindians are a mix of West Eurasians and East Asians. Precisely because it's clear that the Amerindian component back migrated from America.
"The First American component didn't come from East Asians. And it's precisely that First American component that's found in Mal'ta".
Yes, that's what the paper says."
The paper admits logical confusion. On the one hand, it accepts Reich's finding that all Amerindians save Eskimo-Aleuts and Na-Dene are pure in their First American component. It presents new data showing that, in the New World, only Eskimo-Aleuts and Na-Dene are close to East Asians (Han). But then it says that Amerindians descended from East Asians and shortly after their divergence, the First American component entered them from the Mal'ta people.
What would have justified their interpretation if Eskimo-Aleuts and Na-Dene showed proximity to Mal'ta, while the rest of Amerindians showed proximity to East Asians. But this is precisely what we don't see in the data!
It's clear from their ADMIXTURE run, just like from a myriad of others from Rosenberg et al. 2002 on that Amerindians are largely unadmixed and it's precisely this Amerindian component that's found all over Eurasia and even in South Asia and the Middle East. And the Mal'ta boy was very much an Amerindian.
Again, some of the wording in the paper is very unfortunate. We should not forget that both Amerindians and Europeans derive from sister populations of Mal'ta boy - and not his exact group (both his y-DNA and mtDNA are extinct yet derived, and to both sides different haplogroups were present (different U subgroups and IJ and R1 to the west, and Q and X to the East). To both sides the impact of these people - all apparently originating from the NW subcontinent - was actually earlier than Mal'ta boy: the Gravettian started >~32 kya, and Siberia was settled west to east 37ky to 35kya.
ReplyDeleteSo, those ~10ky of drift means that the full contribution of these (outer) populations to both Europeans and Amerindians is likely underestimated in this paper (because it actually derives from populations ~10ky removed from Mal'ta boy).
Mal'ta also had it seems from K=9 more south Asian than European ancestry.
Barak,
Of course - but that just appears like that because you are defining an ancient population via modern components. All Paleolithic and Mesolithic Europeans, northern* West Asians, and non-East-Asian-admixed Siberians very likely originate from the NW part of the subcontinent. Extant Europeans have little S Asian now, because both had 35-45ky to drift apart to make them into exactly these identifiable components now, in the first place.
* Ancient and modern SW Asian may have/ have admixture from Africa and any possible surviving "Arabian Gulf Oasis" population.
The David Reich lab doesn't accept this paper's conclusion that Amerindians are a mix of West Eurasians and East Asians. Precisely because it's clear that the Amerindian component back migrated from America.
ReplyDelete""Mal'ta might be a missing link, a representative of the Asian population that admixed both into Europeans and Native Americans," said Harvard geneticist David Reich. If so, he said, it shows "the value of ancient DNA in peeling back history and resolving mysteries that are difficult to solve using only present day samples."
http://www.upi.com/Science_News/Blog/2013/11/20/Native-Americans-linked-to-ancient-DNA-from-Europe/8931384978750/#ixzz2lLwq9VrR
Couple of comments
ReplyDelete1. There are Siberian populations in the PCA plot who seem to vary on a ML-1 to East Asian axis.
This suggests that they may be more or less admixtures between these two populations (rather than a 3 way East Asian, Amerindian, European combination).
Do we know who they are from the paper?
2. It does seem that this banishes the idea of phenotypically East Asian (or Mongoloid) like admixture in Northern Europe and West Eurasia generally other than is normally historically explained (e.g. Saamis, Turks, etc.).
Instead, the rival hypothesis (suggested back at the time of the first Amerind admix papers) of admixture between a two roughly West Eurasian populations, one of which split off slightly later from Asians and contributed to Siberian (and thereby American) ancestry, rules the day.
The slightly later split of ML-1 from the Asian family, including Onge, Papuan and East Asian family helps explain why there are correlations between f-statistics involving Sardinians, (other Europeans), (Onge / Papuan / East Asian), San. (Although perhaps we still need to be cautious ML-1 was completely unadmixed - as cosmopolitans compared to the other West Eurasian population, they had the opportunity).
But it seems like in terms of actual size, the significant f-statistics found are the showing Amerindian and Siberian like admixture across Northern Europe, and the bulk of derive from shared ancestry from a non-extant, but basically West Eurasian population.
Terry,
ReplyDeleteYour ad hominem usages are getting old.
This is my last statement regarding this particular topic to you:
Basically, what you are calling "Mongoloid," everyone else is calling "Paleo-Mongoloid." And since the accepted Mongoloid genotype and phenotype is based on extant Mongoloids, there is a significant difference between them, making the Paleo-Mongoloid type proto-Mongoloid with respect to extant populations.
I am not making up the concepts of proto-Mongoloid (i.e., the Mongoloid genotype and phenotype feature set present in extreme NE Asia 30-35kya is an identifiable subset of those present in population expansions 10-20kya or later) and the distinction of three different admixture phases in Amerindians - this goes back to decades of archeology, physical anthropology, and genetics. All this should come as no surprise given the drift during that interval (and since then, with respect to extant "extreme" Mongoloid populations).
See old papers such as Williams et al., Am. J. Phys. Anthropol., 66, 1–19, 1985, and Fig. 2 in Reich et al.:
http://www.nature.com/nature/journal/v488/n7411/pdf/nature11258.pdf
The fact that Inuit and Na Dene have a larger Mongoloid genotype and phenotype set therefor is not just because they have more admixture with NE Asian (which is only ~10% in case of the Na Dene - so low it wasn't even picked up in the current paper), but because they have more recent (more Han-like) admixture.
Barakobama and Fanty, both of you misunderstood what I said.
ReplyDeleteI doubt that Y DNA N(specifically N1c) was brought to northeast Europe with Mal'ta like people. I have seen in many other autosomal DNA results that northeast Europeans have obviously modern Siberian ancestry not Mal'ta like. Mal'ta also had it seems from K=9 more south Asian than European ancestry.
I never said that NE Europeans didn't have East Asian-related Siberian ancestry. I said that the populations that brought that ancestry to NE Europe probably also had West Eurasian Siberian ancestry. Imagine Komi-like populations diffusing into Lithuanian-like indigenous Finns rather than a pure Yakut-like population doing the same. This explains the increased signal of Mal'ta-like ancestry in Finns, and also explains why Orcadians are still more Amerindian vs. East Asian than Russians and Estonians etc. It also explains the phenomenon of the disparity between high paternal N and low(er) autosomal East Asian, a contrast most extreme in Lithuanians.
Fanty,
Or the massive Finno Ugric migrations to Britain? (there was no, but still that guy is related to Brits.
But I said that Finno Ugric migrations were possibly a reason why NE Europeans are more related to him, not exclusively related to him. More pre-Neolithic ancestry is also likely a factor. Which factor is more important, I don't know.
This is how I see it:
Native Americans are closest to Mal'ta because they're the descendants either of Mal'ta folk or a very closely related population. Extreme NE Siberians are related to him because, like Amerindians, they're direct or close to direct descendants, but more diluted by subsequent waves of East Asian admixture. Komi, Mari etc. are related because, while perhaps not direct descendants of Mal'ta folk, they're largely composed of similar ancestry (i.e. the Siberian branch of UP West Eurasians). Britons are related because they derive a large portion of ancestry from the related European branch of UP West Eurasians. NE Europeans are the most related Europeans because they have more UP European + UP Siberian ancestry, but also East Asian, which affects the asymmetry on the test I talked about above.
I recall former experiments to calculate the timespan of the admixture event.
those calculations said, Amerindian admixture in northern Europeans is from 17K years ago and exists in all Europeans, in northern Europeans twice as much as in southern ones while there is only a tiny difference between Irish and Russian.
The same calculator said, East Asian admixture in North-East Europeans is from 2500 years ago and exists in Russians but zero in Irish.
I know the Irish don't have any East Asian admixture. In fact, I've been arguing all along that in most Europeans the Amerindian signal is unrelated to East Asian admixture. I've never seen that 17,000 ya estimate of Amerindian admixture in Europe, so I'm not sure what assumptions were involved in its calculation.
When I look at K = 9 map, southern Native Americans do not have any yellow or light yellow colour (East Asian, Siberian). According to Fanty, ”those calculations said, Amerindian admixture in northern Europeans is from 17K years ago and exists in all Europeans, in northern Europeans twice as much as in southern ones while there is only a tiny difference between Irish and Russian.” If you say that the Amerind admixture (orange colour) in European and North Eurasian populations dates from 17k years ago, how is it possible that Amerinds would be predominantly East Asian like, if there is no East Asian admixture components in them (both northern and southern components are lacking)?
ReplyDeleteEurologist said in a previous thread that ”also, in such an old population, tools like Admixture will always make them look pure and unadmixed - that carries little meaning by itself, because we have too few ancient reference populations (ancient DNA).” If this is true, is it possible that the Amerind admixture in Europeans is as old as 17K.
Basically I would like someone to explain me why the ancient 15k Amerind admixture in Europeans is visible but the ancient massive East Asian admixture in Amerinds is not?
Yes. This is the official statement, which is already different from Raghavan's conclusion as it suggests that gene flow went into West Eurasians as well, not just into Amerindians. This is half way toward a back migration idea.
ReplyDeleteAnd here's what an e-mail to me says:
"I think you'll be pleased to know that I do consider quite interesting and worthy of more thorough investigation indications of back-migration or complex bidirectional interaction between the New World and Eurasia, and believe that contemporary and ancient American genomic data may well play a useful role in disentangling archaic/modern interactions. I will also say that our group does have some reservations about the scenario Eske proposed.., given the potentially confounding factor of Amerindian-related North Asian gene flow into prehistoric Europe (as outlined in Nick Patterson's 2012 "Ancient Admixture in Human History"), which prehistoric West Eurasia > Americas input is by itself clearly insufficient to explain."
@ Eurologist.
ReplyDeleteI think the 4% Papuan is being lost in discussions between South Asian, Euro and Amerindian components. This has to be tied in with the South Asian (which peaks in the Sakkiliar who are a south Indian group). Hence the admixture looks "like" ASI. Look at the Harappaworld spread sheet for any of the South India populations and you will see what I am talking about. The trace (but non-significant) Papuan shows up all over the place.
Nut you are correct. One cannot derive ancestry based on components that have drifted into different populations today.
@Kristiina
ReplyDelete"Basically I would like someone to explain me why the ancient 15k Amerind admixture in Europeans is visible but the ancient massive East Asian admixture in Amerinds is not?"
Finally, the voice of reason in a jungle of ignorance and superstitions. Kristiina, you are getting my Web Gem Reward this week.
Dienekes does the study mention the pigmentation?
ReplyDeleteYou posted a tweet about it but no excerpt on the release study conclusion here on that. Some news report has this:
"The first is that the boy's DNA matches that of Western Europeans, showing that during the last Ice Age people from Europe had reached farther east across Eurasia than previously supposed. Although none of the Mal'ta boy's skin or hair survive, his genes suggest he would have had brown hair, brown eyes and freckled skin."
http://www.ndtv.com/article/world/two-surprises-in-ancient-dna-of-boy-found-buried-in-siberia-449216
Matt,
ReplyDelete1. There are Siberian populations in the PCA plot who seem to vary on a ML-1 to East Asian axis.
This suggests that they may be more or less admixtures between these two populations (rather than a 3 way East Asian, Amerindian, European combination).
Do we know who they are from the paper?
The supplementary data is open access and contains some things you may find interesting. Basically, the relationship of Siberians to Mal'ta is roughly proportionate to their West Eurasian ancestry (component: European) on Dienekes's calculator here:
https://docs.google.com/spreadsheet/ccc?key=0ArJDEoCgzRKedGR2ZWRoQ0VaWTc0dlV1cHh4ZUNJRUE#gid=0
Yakut, Buryat and Mongola are among the less related, while Ket and Khanty are among the most. All Siberian groups at k=3 have some degree of 'European' ancestry, with most seeming to have around 10%, and these seem to constitute the central bulk of Siberians in their relation to Mal'ta (with some fairly minor exceptions), and the East Asian groups that seem to share some of the Mal'ta drift seem also to be the populations with some minor European at k=3 vs. the less admixed East Asians.
2. It does seem that this banishes the idea of phenotypically East Asian (or Mongoloid) like admixture in Northern Europe and West Eurasia generally other than is normally historically explained (e.g. Saamis, Turks, etc.).
Yes, in the paper Mal'ta's East Eurasian affinities were tested. He was more related to Papuans than to Han. The percentage wasn't quantified, but from the figure his Papuan vs. Han affinities were about the same as in the modern SW Asian populations included (which is likely ASI related in the latter, or related to some third variable such as SSA admixture). Most of the other West Eurasian populations included hovered around the middle neither-nor point, or were shifted slightly (or strongly in the case of Russians, Adygei and some S. Asians) toward Han.
Willerslev suggests minor admixture with a local population or errors resulting from lower quality DNA.
Dienekes does the study mention the pigmentation?
ReplyDeleteLook at the very end of the freely available supplement.
Matt, further to that last comment, check out figure SI 21 here:
ReplyDeletehttp://www.nature.com/nature/journal/vaop/ncurrent/extref/nature12736-s1.pdf
It's not the data for the PCA plot you were referring to, but it explains the pattern adequately. Basically, some Siberians share considerable drift with Mal'ta, as much as, or more than, some Europeans, but as we see from the PCA plot they are further from Mal'ta than Europeans. This suggests to me that while Mal'ta is extremely closely related to a portion of the ancestry of Siberians, the majority ancestry in Siberians forces them much further from Mal'ta than the Europeans are (who have more compatible majority ancestry with Mal'ta), because the East Asian portion of Siberian ancestry has relatively no relation.
So it seems to be that East Asian migrants into Siberia 'mopped up', if you will, extremely close lineages to Mal'ta, which seem to constitute a large portion of their West Eurasian ancestry, and the relationship of this ancestry with Amerindians, in addition to these tribes' close relationship with the East Asian portion of Amerindian ancestry also, gives them an Amerindian signal that isn't actually real admixture. I think this explains the PCA.
It also seems from the same figure that Lithuanians share more drift with Mal'ta than Maris do, so for now I take back my comments of Mari/Komi-like ancestry bolstering the relationship between NE Europeans and Mal'ta pending further evidence (though according to Vadim's calculators, mal'ta still seems to be most related to Udmurts).
Sorry Matt, I see now you were referring specifically to the East Asian to MA-1 limb of the Siberian triangle formation.
ReplyDeleteThe answer to your question is here:
http://www.nature.com/nature/journal/vaop/ncurrent/extref/nature12736-s1.pdf
Figure SI 10
I've not had time yet to look into what distinguishes the two strands of Siberian, but looking at a few k=4 values, it seems the Siberians streaming in the direction of Amerindians have much higher levels of Amerindian. I'll have a think about it.
OK thanks, Dienekes.
ReplyDeleteDoes anyone actually remember how I argued that ANI-West Asian-North European split somewhere in Iran or Central Asia and moved into different directions.
ReplyDeleteI believe before these three split they had even a common origin with East Eurasians.
"People follow the opinions of authoritative others. They don't look at the data".
ReplyDeleteHamarfox, Lathdrinor and I have each based our conclusions only on the data. The difference between us and others is that we accept the data, unlike many others who have a preconceived idea of what the data should show.
"What would have justified their interpretation if Eskimo-Aleuts and Na-Dene showed proximity to Mal'ta, while the rest of Amerindians showed proximity to East Asians. But this is precisely what we don't see in the data!"
That surely is the point. The data show that Eskimo-Aleuts and Na-Dene are less related to the Mal'ta boy than are the remainder of Native Americans. That is why they can come to the conclusion they do. We can assume that Eskimo-Aleuts and Na-Dene represent a less-admixed population than do the bulk of the Native Americans.
"It's clear from their ADMIXTURE run, just like from a myriad of others from Rosenberg et al. 2002 on that Amerindians are largely unadmixed"
That is because until now they have been assumed to be representative of unadmixed Mongoloids. We now see that this assumption was wrong and so the relationship needs to be looked at again.
"the Mal'ta boy was very much an Amerindian".
No he wasn't. Native Americans have just '14 to 38%' in common with the boy.
"Extant Europeans have little S Asian now, because both had 35-45ky to drift apart to make them into exactly these identifiable components now"
And any number of individual admixture events.
"Basically, what you are calling 'Mongoloid,' everyone else is calling 'Paleo-Mongoloid'."
'Everyone else'?
"since the accepted Mongoloid genotype and phenotype is based on extant Mongoloids"
And 'extant Mongoloids' include a variety of populations that have undergone a variety of admixture events as the Mongoloid phenotype expanded. 'Everyone else' certainly accepts that to be so in South China, Southeast Asia and the Pacific. What the present paper shows is that the same holds for at least the Native American portion of the northern Mongoloids.
"I never said that NE Europeans didn't have East Asian-related Siberian ancestry. I said that the populations that brought that ancestry to NE Europe probably also had West Eurasian Siberian ancestry ... It also explains the phenomenon of the disparity between high paternal N and low(er) autosomal East Asian".
Including the relative lack of the EDAR370A mutation, where we would expect to find it from the presence of Y-DNA N.
My mother (Irish) has Amerindian DNA segments which are specifically found in Athabaskans and she also has Chukchi DNA segments, and about 4% Caucasus (Lezgin and/or Nogay) - so this includes the entire Dene-Yensieian-Caucasian range.
ReplyDeleteOn Marnie's blog at the beginning of the year, I predicted:
http://linearpopulationmodel.blogspot.com/2012/12/little-big-man.html?showComment=1357255570559#c960607464383659173
"My thinking is that the Salishan-Irish connection would have to be a shared Central/Northern Asian connection from the Mesolithic - like that old-style singer video I posted previously."
Also here:
http://linearpopulationmodel.blogspot.com/2013/01/on-origin-of-salish-wakashnan-and-north.html?showComment=1359147621313#c1027547612377599963
Also:
Have you seen the new DNA Tribes paper suggesting that one of the formative peoples of the current European population is none other than the SALISH/SALISHAN peoples?!?!?!?!
http://www.dnatribes.com/dnatribes-digest-2013-01-02.pdf
The Salishan component is estimated at 5.5% of their "Celtic Tribe" - which is centered in Ireland and Western Britain - the highest in Western Europe, with only the "Urals Tribe" having higher at 6.5%
In term of what these mystery people - Old Siberians or Hyperborains looked like, as I posted previously, I'd think something like this Irish singer from the West of Ireland, who has a vaguely Amerindian cast to his features:
ReplyDeletehttp://www.youtube.com/watch?v=vGPUO1yEE7Y
Check for a close up at 0:12 to 0:22
Of course this is also a Cro-Magnon like look also?!
Keep in mind that Lake Baikal was a fresh water lake that likely didn't freeze during the Ice-Ages. If the Tocharians are considered, the child could have been a descendant of populations living near the life-providing Baikal long before migrations to the Americas. In other words, Asians and Europeans camped at Lake Baikal together until the ice melted. Not much different than the Xiangno-Tocharians in Xinjiang today I bet.
ReplyDelete@Kristiina
ReplyDelete"Basically I would like someone to explain me why the ancient 15k Amerind admixture in Europeans is visible but the ancient massive East Asian admixture in Amerinds is not?"
Because the Amerindian have a basic root in paleolitics of tropical America, where they survived the larger part of ice-time. The 220.000 yrs old Hyatelco is ewvidently from an archaic human that no-one seems to understand have existed.
Seemingly there were people in Europe who dared to escape the beginning of the European Glacial Maximum (17-21.000 yrs BP) by following the Gulf-stream west of Shetland to the coast of N-Amerika.
We know that the Cro-Magnon occupying NW as well as NE Europe during paleolitiukum had mt-dna U/U5, T and X. Today - after the Malta boys testimony - we also know that they had y-dna R. That implies that the entire macrogroup F are designed by an paleoarctic population in northern Eurasia.
The best chance such a population would have to survive the very roughest of cold-periods, such as Dryas I and III, would be the atlantic facade, presumebly where a steady gulf-stream could give an optimal effect.
To find the last and final refugia of the the Solutrean/Hamburgian haplogroup U/T and I/R that remained in Europe AND were able to survive the Last Glacial Maximum - we have to remain by the Atlantic facade.
During this stage the last mammuts died away - in present Denmark, Skania and Estonia - while all the rest of northern Eurasia were merely tundra and taiga.
Obviously a very limited number of hgumans managed to survive this period - in an NW refugia. This "bottleneck" is by now a well known phenomenon - and slogan - within population genetics.
By then some of the Solutreans had created the pre-Clovis societies in north-east America - where the tropical, indigenoius ameazonians hardly cared to live...
As soon as ice-time was over we had an immediate re-colonisation of the entire northern Eurasia - by new descendansts of the very same NW Cro-Magnid haplogroups that several generations before had to leave Denisova, Sungir, Kostenki and Dolni Velistoce.
Their refugia is still unknown, but they must have survived somewhere close to Ahrensburg. The last N Eurasian population known before Younger Dryas point to Ahrensburg at 12.700 yr BP. During the "big chill" (12.700 - 12.300 BP) all traces of people within northern Eurasia disapear - before the first known mesolithics re-apear, again at Ahrensburg, around 12.200 BP.
Repopulating northern Eurasia the paleolithic haplogroups - such as hg U and R - could again get in touch with east and south-east Asians - and mix with proto-mongoliians/chineese.
The result came to be the modern NE Asians and Tibetans. Theesenew NE asians cicumvened the Arctic Ocean, eastwards to Alaska and Greenland - westwards to the Kolan Penninsula and northern Fenno-Scandia.
Consequently we may find specifically NE Asian genes in the NW America, while the old European haplogroups – such as ydna R and mtdna X - concentrated in NE America - exactly where the Pre-Clovis/Solutrean artefacts are found.
Maris have a high frequency of R lines, 29% of R1a, 10% of R1b, while Udmurts have considerably more N1c (58%) and less R1a (10%). According to my old papers, Udmurts also carry a high frequency of K, of which I would really like to know what it is. If the Udmurts are the closest match to the Mal’ta boy, we should take into account their main haplogroups:
ReplyDeleteN1c 58%, K 25%, R1a 10%
T 22%, H 21%, D 12% U2 10%, U5 9%
Terry, I did some photo googling with “Nenets”, and I do not think that N folks originally carried EDAR thick hair variant. There was one photo that really impressed me (http://stephandudeck.files.wordpress.com/2012/05/img_1781.jpg), because the younger woman in the photo looks very much like my relatives from my father’s side, including the hair structure and colours. Also her eyes look just the same as the eyes of many of my aunts and uncles! However, with this, I am not saying that she is representative of the original N folks.
I am not sure about the route of N1c to Europe, but please note that there is no northern yDNA path from Northern Siberia to Europe, i.e. European N lines have not developed from Yakut, Nenets, Nganasan or even North Chinese lines. According to this map http://rokus01.files.wordpress.com/2010/03/bashkir_5.jpg, the root of N1c is shared between West Siberians, Volga-Ural people and South Siberians and Central Asian people.
As after the Last Glacial Maximum Europe was recolonized from the Middle East, doesn't this suggest that one likely possibility is that we are talking about a group from Middle East that went west into Europe and East into Siberia and ultimately North America? So no European roots?
ReplyDeleteSorry for doubleposting! Have a look at Khanty Alyona Nemelkina http://www.nba.fi/hanti/en/a4_s1.php
ReplyDeleteI would bet that she has EDAR370A. You must also remember that Northern Khantys have assimilated a remarkable number of Kets, the frequency of yDNA Q-L330 is 21%. This is just one of the lines that according to German has backmigrated to Siberia.
"Your ad hominem usages are getting old".
ReplyDeleteSorry Eurologist (and German), but I get very frustrated when others can't see what I find so blindingly obvious. Especially when that inability to see arises through commitment to a belief that requires ignoring so much other data.
The current paper shows (to my satisfaction anyway) that Native Americans cannot be considered 'proto-' or 'paleo-Mongoloids' in any way. They are a hybrid between a completely non-Mongoloid Central/West Eurasian population (represented by the Mal'ta boy) and an East Asian population. In fact this paper merely reinforces what seemed obvious in this 1999 paper:
http://ac.els-cdn.com/S0002929707617200/1-s2.0-S0002929707617200-main.pdf?_tid=3a30027e-5481-11e3-b161-00000aab0f26&acdnat=1385240155_849e437644380347c2f46894e6b56bca
In contrast to the source population of Native American Y-Chromosome haplotypes, quote:
"The scarcity of the NewWorld founding mtDNA haplogroups in Asian populations has also been used as evidence against multiple migrations from different source populations, whereas their circumscribed presence in Mongolian, Tibetan, Korean, and Taiwanese/Han Chinese populations has been used to support the
theory of a single migration of NewWorld mtDNA lineages from Central Asia (Merriwether et al. 1995)".
Other data available shows that the East Asian element involved in Native American ancestry was certainly homozygous for the EDAR370A mutation, which is especially common in Americans, according to the paper:
http://www.picb.ac.cn/picb-dynamic/admin/pic/Cell-WSJ.pdf
The gene's geographic origin lies between about 32-38 degrees north and 110-115 degrees east. Kristiina reminded us that the Tianyuan skeleton, found within this distribution, was Mongoloid. It is therefore reasonable to suppose that by the time of the hybridization event the East Asian element did not contain only the EDAR element of the Mongoloid population and was already 'fully-Mongoloid'. So not even the East Asian element of the admixture should be considered 'proto-Mongoloid'.
Dienekes, a bit off topic, but would it be possible that you make a analysis of pigmentation with only a set of pigmentation SNPs with the sample that you have with Dodecad Project ? Thanks,
ReplyDelete@TerryT
ReplyDelete"Hamarfox, Lathdrinor and I have each based our conclusions only on the data. The difference between us and others is that we accept the data, unlike many others who have a preconceived idea of what the data should show. "
I forgot to mention that one needs to be appropriately trained to be able to look at the data and make conclusions. You are not trained, Terry, that's why you see what doesn't exist. And if you don't have a theory to explain facts, you shouldn't be in the business of making conclusions.
"We can assume that Eskimo-Aleuts and Na-Dene represent a less-admixed population than do the bulk of the Native Americans."
You can assume, Terry, not we. There's nothing to assume. Look at the ADMIXTURE plot and you'll see that Na-Dene and Eskimo-Aleuts carry more ancestral components than Karitiana and Surui.
"That is because until now they have been assumed to be representative of unadmixed Mongoloids. "
It's not the purity of Amerindians that's an issue. It's their derivation from Mongoloids. The data shows no connection to Mongoloids (look at the yellow and light yellow) and reaffirms the purity of - at least - South Amerindians.
"No he wasn't. Native Americans have just '14 to 38%' in common with the boy."
That's what I was referring to.
"I would think, Y-DNA from a >20K year old corpse would have been declared "absolutely impossible" only 5 years ago."
ReplyDeleteWell, recently they managed to sequence the genome of a horse that lived over a half million years ago.
I could agree on "Proto Mongoloid" being a misnomer that merely identifies possibly admixed populations in the periphery of a more local, possibly archaic influence. I'd rather assume the true Mongoloid source was a hybridization event that involved intrusive non-mongoloid elements like also found in the Zhoukoudian Upper Cave near Peking. E.g., the orbits of the Old Man of Upper Cave 101 are relatively low and rectangular, not unlike the European Cro Magnon 1 skull that is about the age of Mal'ta boy. Instead, living East Asians and Native Americans have a facial skeleton characterised by great facial height and high orbits. Kamminga (1992) argued this specimen to be distinct from the “modern Mongoloid morphology” and to be relatively close to that of the Ainu - generally considered physically similar to Caucasians. If so, this apparent ancient European signature of Ainu has by now been obfuscated by "more recent" expansions of East Asians - that IMO actually consisted of a new brand of hybrids. However, since we don't know the genetic composition of the archaic source population (~ Ordos Man from Salawusu site?), it remains impossible per definition to establish the admixture components of such hybrid East Asians in relation with modern human immigrants like Mal'ta boy. In other words, the apparent absence of an East Asian component in Mal'ta boy and vice versa wouldn't have any meaning if East Asians were truly hybrids. And yes, now I insinuate European influence in the formation of the East Asian type.
ReplyDeleteThe main riddle now seems to be the Amerindian affinity of Mal'ta boy MA-1 without East Asian admixture, while Amerindians also diverged from East Asians. This implies an Amerindian component that is separate from the East Asian group. However, is this element just a product of geographic distance from Europe or due to yet another local archaic or "Beringian" source? I gather this to be the quirky outcome of early hybridization with a surplus of some kind of differentiated European influence compared with their East Asian brethren.
Then, the same hybrid lineage at the origin of East Asians was also involved in the peopling of the Americas. The Amerindian "purity" may very well be acquired later on since Figure SI 24 (Test for differences between 51 Native American populations in their affinity to MA-1 compared to the Karitiana) still shows some remarkable fluctuations into an East Asian direction e.g. for Huatar (Chibchas in Costa Rica) and Yaqui (Uto-Aztecan population in Northern Mexico), in a single wave of "First Americans" or by strong dilution of a secondary immigration wave of more East Asian signature.
Gene flow of the Mal'ta boy type also returned to Europe, as has been shown for Northern Europeans. Amerindian background values for other Caucasian populations can now be discarded as ancient relatedness. I hold La Brana, apparently kept outside the Amerindian gene flow back into their northern kin, as the ultimate proof of this formative division. So far I hold any other possible Amerindian migration back into Eurasia to be specifically Na-Dene and more recent.
BTW. I don't think my hypothesized archaic Mongoloid component was closely related to Denisovans, but maybe some Mal'ta-like immigrants (or kin of their ancestors) absorbed their genes and moved on to follow a coastal route to Oceania. At least the closer genetic affinity of Oceania with Europe compared to East Asians can be observed in Raghavan's figure 1b.
Hamarfox, thanks for your responses.
ReplyDeleteOn German's discussion of components, it might be useful to compare the statements by Eurogenes / Davidski on the use of ADMIXTURE in this paper with German's (quoted bits Eurogenes) -
"The reason for these surprising results" (where ancient samples are split between modern clusters) "is that there aren't enough ancient genomes yet to create ancient clusters. When these ancient clusters start appearing, the hunter-gatherers will stop looking like mixtures of modern clusters, and modern genomes will start looking like mixtures of ancient clusters. I explained this in my other blog entry."
and
"You don't understand. It doesn't matter if the ancestral alleles of modern Amerindians originated in a European-like population or not. Today they're typically Amerindian, and seen as such by the software, especially because Amerindians have low haplotype diversity due to multiple founder effects and drift.
In other words, it's easier for Amerindians to create ancestral populations or components specific to them, and the model-based software is more likely to see the ancient links between Amerindians and Europeans as gene flow from the former to the latter."
Davidski's comments seem to me to pretty obvious to anyone who has a basic understanding of how this cluster analysis works and has kept up to date with even the dumbed down blog post regurgitation of work on admixture in ancient populations. For all that German obviously has lots of technical knowledge on kinship structures, and is an expert on this, and that is very useful and interesting knowledge and a good reason for him to be along at these conferences and publish books and so on...
Here you have an exciting photo of Kets taken by the Norwegian explorer Fridtjof Nansen in the beginning of 20th century http://img14.imageshack.us/img14/6920/ket4.png
ReplyDeleteI would say that there is definitely something Native American about them.
Basically I would like someone to explain me why the ancient 15k Amerind admixture in Europeans is visible but the ancient massive East Asian admixture in Amerinds is not?
ReplyDeleteKristiina,
If you take a look at the PC plots, you can see that most "First Americans" are hugely drifted away from other populations. That's why they form their own cluster so early (K=4). According to one study, it was estimated that they spent ~20,000 years in almost isolation in Beringia before even moving into the Americas. At any rate, it is clear that the admixture between West Eurasian-derived Paleo-Siberians and Paleo-E Asians took place a long time ago, so current populations are not a great match for that (since they have drifted, since then), while Amerindians have both homogenized and also drifted (in the PC1/2 plot, many "First Americans" are further away from central populations than either Europeans or E Asians, likely reflecting the small, random subset of SNPs of the Paleo-W/C Eurasian founder population, a drift of that population, and a small subset of paleo-E Asian SNPs in their founder population, and subsequent drift over 20-35ky).
One thing one could do to address your question (that I would find very interesting), is to omit Amerindians from the ADMIXTURE calculation and then see how the other populations project into them. One could do a run with and without the most eastern (or most Han-like) Siberians, to double-check which population is closer but also to avoid that one becoming the default fall-back population. I don't know if any of the Amerindian data are public domain - then Dienekes (or anyone else) could do such a calculation.
And again, I think the "majority opinion" now seems to be that the Amerindian signature in Europeans is largely caused by the commonness of Paleo-Siberians moving both West and East (just before the Gravettian), and not by East-Asian admixture.
PC3 is pretty much useless since it is determined by Oceania, here, which clearly should have been omitted. PC4 largely shows E Siberian and Inuit drift - which have drifted away from all other populations, except MA-1 already shows a bit of that. Na-Dene also does but Karitiana doesn't, so my interpretation is that this is a portion of the Mal'ta boy's genome that wasn't present in the Paleo-extreme E Siberians that actually mixed and formed First Americans, but was present slightly further West and has survived in extant Siberians and in more recently-admixed Na-Dene.
@Matt
ReplyDelete"Davidski's comments seem to me to pretty obvious to anyone who has a basic understanding of how this cluster analysis works"
What Davidski wrote is fine and dandy but the very fact that Amerinians have "low haplotype diversity due to multiple founder effects and drift" makes them a perfect model of ancient Late Pleistocene and even Mid-Pleistocene populations. Don't assume that it's a product of recent population fragmentation brought about by the "peopling of the Americas" in the past 12-15,000 years. The latter time frame is not based on any facts - genetic or archaeological - and hence to be proved and not assumed.
At least two Amerindian components (PINK and ORANGE) seem to have migrated out of America, with no YELLOW (Asian) components entering it. What ADMIXTURE can't detect if whether the West Eurasian (BLUE) component entered the New World and admixed with the other two or the New World is the place where it originated.
Also compare the Mal'ta ADMIXTURE plot with the early cluster analysis by Rosenberg et al. 2002 (http://anthropogenesis.kinshipstudies.org/2013/11/ancient-dna-from-malta-and-afontova-gora-a-full-account/) which I listened to as a part of the Marcus Feldman seminar at Stanford before it was actually published. You'll see that at K = 2 the whole world is divided between an African and an Amerindian component, with the latter detectable at low frequency in Sub-Saharan Africa as well.
"German obviously has lots of technical knowledge on kinship structures, and is an expert on this,"
The population genetics of modern humans is either part of anthropology (to which kinship studies and reflexive thinking are core) or it's nothing.
@Rokus
ReplyDelete"Instead, living East Asians and Native Americans have a facial skeleton characterised by great facial height and high orbits."
The earliest skulls with "Mongoloid" morphology come from the Americas, not from Asia.
"This implies an Amerindian component that is separate from the East Asian group. "
Two components to be precise - ORANGE and PINK. Both in Mal'ta, both all over modern Eurasians. The PINK one fits the Na-Dene migration back and the appearance of Kets in SIberia.
"The Amerindian "purity" may very well be acquired later on since Figure SI 24 (Test for differences between 51 Native American populations in their affinity to MA-1 compared to the Karitiana) still shows some remarkable fluctuations into an East Asian direction e.g. for Huatar (Chibchas in Costa Rica) and Yaqui (Uto-Aztecan population in Northern Mexico)."
It is indeed interesting. Good catch! This is exactly what we see in craniology. On the other hand, Tianyuan is deemed ancestral to modern East Asians and the population, which is closest to it, is Karitiana (http://anthropogenesis.kinshipstudies.org/2013/11/ancient-dna-from-malta-and-afontova-gora-a-full-account/). So, whether we take West Eurasians or East Asians, the range of variation found in the Americas (also supported by ancient and modern craniology) can accommodate a founding migration into any part of Eurasia - eastern and coastal or western and inland.
This discovery somehow strengthens my suspicion that what we call the "Classic Mongoloid type" came from a founding population somewhere around, perhaps, Sikhote-Alin? Their traits and some related genes made it into late Dyuktai and eventually to Eskimo-Aleut. But the great profusion of the particular package of traits and genes we call "East Asia" comes from a Neolithic population expansion starting around 8,000 B.C. (rice, millet, pottery)
ReplyDeleteThis comment has been removed by the author.
ReplyDeleteThe Malta sample has U as most mt-dna found in the paleolithics of Eurasia. Consequently he's from the same base as all other paleolithic and mesolithic mt-U found in Eurasia - from Span and France to Ukraine and Siberia.
ReplyDeleteMoreover he's y-R, from the same archaic cluster as the earliest mesolitics.
Thus he's "typical" west Eurasian, or what? Can someone be kind enough to explain why the affinities between these NW h-groups and the amerindian genome can't be the result of an EUROPEAN migration during the late paleolithic, such as the Solutreans and/or Magdalenians.
Moreover - how can geneticans exclude the Mid-Paleolitic Hyatelco-culture as a factor to recon with?
There's still an emperor out there somewhere - without clothes.
And here's what an e-mail to me says:
ReplyDelete"I think you'll be pleased to know that I do consider quite interesting and worthy of more thorough investigation indications of back-migration or complex bidirectional interaction between the New World and Eurasia, and believe that contemporary and ancient American genomic data may well play a useful role in disentangling archaic/modern interactions. I will also say that our group does have some reservations about the scenario Eske proposed.., given the potentially confounding factor of Amerindian-related North Asian gene flow into prehistoric Europe (as outlined in Nick Patterson's 2012 "Ancient Admixture in Human History"), which prehistoric West Eurasia > Americas input is by itself clearly insufficient to explain."
German, whose e-mail is this?
There is yet another possibility for the vector of connection between Mal'ta Boy and NW Europe - that is a reverse Solutrean... or early Native Americans, in the NE of the continent making a North Atlantic crossing to NW Europe after the Ice finally cleared from there.
ReplyDeleteHere are my mother's Eurogenes K13.
Population
North_Atlantic 49.29%
Baltic 26.58%
West_Med 12.76%
West_Asian 6.07%
East_Med 0.69%
Red_Sea 1.18%
South_Asian 0.19%
East_Asian -
Siberian -
Amerindian 1.39%
Oceanian 0.78%
Northeast_African 1.07%
Sub-Saharan -
On this test she has Amerindian, but no Siberian or East Asian.
There is also the similarity of the word for hide/skin boat in each case:
1. Irish - Currach - pronounced KURR-OK
2. Native American - Kayak - pronounced KAI-YAK
words similar except for some vowel changes.
Hey Dienekes, here are their genomes
ReplyDeletehttp://www.cbs.dtu.dk/suppl/malta/
"You are not trained, Terry, that's why you see what doesn't exist".
ReplyDeleteI guarantee I have had more experience with pratical genetics in all its forms than you have.
"When these ancient clusters start appearing, the hunter-gatherers will stop looking like mixtures of modern clusters, and modern genomes will start looking like mixtures of ancient clusters".
And that whole pattern of movement and mixing goes way back:
http://dienekes.blogspot.co.nz/2013/10/d4500-and-unity-of-early-homo.html
"this apparent ancient European signature of Ainu has by now been obfuscated by 'more recent' expansions of East Asians - that IMO actually consisted of a new brand of hybrids".
I basically agree. But I see the hybriization events that gave rise to the Mongoloid phenotype occurred at least 30,000 years ago.
"I think the 'majority opinion' now seems to be that the Amerindian signature in Europeans is largely caused by the commonness of Paleo-Siberians moving both West and East (just before the Gravettian), and not by East-Asian admixture".
Yes.
"This discovery somehow strengthens my suspicion that what we call the 'Classic Mongoloid type' came from a founding population somewhere around, perhaps, Sikhote-Alin?"
I think we can place it fairly securely in the region I mentioned above: between about 32-38 degrees north and 110-115 degrees east.
""we don't know the genetic composition of the archaic source population (~ Ordos Man from Salawusu site?)"
We have a pretty good idea of where it originated though. Somewhat west of Beijing for a start.
"I do not think that N folks originally carried EDAR thick hair variant. There was one photo that really impressed me"
But the Nenets are a fairly westerly group of N.
"I am not sure about the route of N1c to Europe, but please note that there is no northern yDNA path from Northern Siberia to Europe"
Which gives plenty of opportunity for their ancestors to have mixed considerably with non-EDAR people.
"And yes, now I insinuate European influence in the formation of the East Asian type".
Not necessarily 'European' but something similar to the Mal'ta
population. But I think such admixture is later than the event that was involved with the Native Americans. In fact the 'original' Mongoloid population probably doesn't exist any more. It has been diluted by later population movements.
"I don't think my hypothesized archaic Mongoloid component was closely related to Denisovans, but maybe some Mal'ta-like immigrants (or kin of their ancestors) absorbed their genes and moved on to follow a coastal route to Oceania".
I mostly agree but I would guess that the population carrying the Denisova element via the coastal route to Australia/New Guinea was much earlier than the Mal'ta population's arrival, and missed the ancestors of the Mongoloids completely.
@batman: What is this "Mid-Paleolitic Hyatelco-culture"? I can't seem to find anything on google.
ReplyDeleteAs for my own previous comment, in case it was confusing, I was mainly just speculating that perhaps the reason East Asians aren't particularly close to the boy from Malta is, their genetic profile comes from a later founding population that expanded with the Neolithic.
What would be helpful in untangling this issue of the relationship of Native Americans to Eurasian populations would be more genetic testing. Geneticists have lamented the lack of cooperation in DNA testing of native populations of the United States. Indeed, DNA testing of Native Americans has become highly politicized. The Beringian Model has unfortunately become dogma and has resulted in genetics researchers adopting tunnel vision. Now, many Native Americans are afraid to be tested for fear they might lose their tribal memberships if they test for haplogroups not generally recognized as “Native American”.
ReplyDeleteThere may be actual differences between the eastern and western native populations of North America that could reflect different origins. The western populations, which also gave rise to the Meso and South Americans populations, are obviously derived from the Beringian population and reflect an admixture of Siberian moving east from the interior and East Asian populations moving up the Pacific Coast. The eastern branch, on the other hand, may have originated from deeper in Siberia and may have more closely resembled the Mal’Ta population. There have been some controversial results of DNA testing on Cherokee women that purports to show a large proportion of U5 (that is, the Mal’ta boy’s mtDNA type) as well as T, but very little H – in other words, approximating the model of European hunter-gatherers. The presence of mtDNA X is also a strong indication of real differences between the eastern and western Native American populations. The problem here is that we are having to try to determine the original composition of a population that has undergone a recent admixture with another population to which, as we have noted, is already somehow related to it. It is something like trying to read the original text of a parchment that had been scraped off and overwritten.
Based on my linguistic analysis as well as DNA distributions, I would suggest that the Greenberg’s Three Wave model of the settling of the Americas be modified to include the Algonquian and Iroquoian –speaking populations as well as the Na Dene. This “second wave” would appear to have a composition more reflective of ancient interior Siberian populations. More DNA testing needs to be done on these populations with mind open to accepting the possibility that there may have been other founding lineages in addition to the standard model A, B, C, D, and X mtDNA groups as well as YH Q and C.
It is worth noting that there appears to be support for the view that Basque shares a deep affinity with Na Dene language group. For instance, the Basque word for “water” – ur – can be linked to the Ket equivalent -ul - by the simple r -> l phonetic shift observed in other languages. Surprisingly, however, I also found a lot of points of similarity with Algonquian and Iroquoian languages in the pronoun system, which may have some bearing on the question of the relationship of early European and North American populations. The Basque population is overwhelming HG R1b, and share a subclade with Bashkir. Analysis of R1b subclades among Native Americans might be fruitful.
Not necessarily 'European' but something similar to the Mal'ta
ReplyDeletepopulation. But I think such admixture is later than the event that was involved with the Native Americans. In fact the 'original' Mongoloid population probably doesn't exist any more. It has been diluted by later population movements.
At least EDAR370A indicates the original Mogoloid population was the result of a unique hybridization event. The archaic genes may have diluted, but not the hybrid genes that became fixed in the East Asian type.
I would guess that the population carrying the Denisova element via the coastal route to Australia/New Guinea was much earlier than the Mal'ta population's arrival, and missed the ancestors of the Mongoloids completely.
We don't know when the ancestors of the Mal'ta population arrived in the neighborhood, the genetic data and anthropological evidence I mentioned suggest they were part of a much wider and older movement, even compared to the Amerindian divergence.
According to Rokus, ”I don't think my hypothesized archaic Mongoloid component was closely related to Denisovans, but maybe some Mal'ta-like immigrants (or kin of their ancestors) absorbed their genes and moved on to follow a coastal route to Oceania. At least the closer genetic affinity of Oceania with Europe compared to East Asians can be observed in Raghavan's figure 1b.”
ReplyDeleteAccording to German, the Mal’ta sequence (MA-1) occupies its own clade within hg U. But what transpires if one peels the onion on it and compares all these clade and sub-clade defining mutations with the existing ancient mtDNA from extinct Eurasian hominins (Denisovans and Neandertals) is that 20%+ of these major substitutions defining the internal structure of the U clade in modern humans have exact matches with Denisovan substitutions. On MA-1 sites 13350 and 16399 are such “ancestral retentions.” The latter one is polymorphic on Denisovans, which is a rare case as Denisovan mtDNAs from the tooth and from the pinkie are nearly identical. … Considering that these are direct matches with the hominin DNA collected in the same general area of South Siberia as Mal’ta DNA is from, I find it pretty interesting in the light of the recent discussions I had about “archaic admixture” on mtDNA.”
According to the supplementary material of this Mal’ta paper, “we wanted to address the curious finding that TreeMix identifies an admixture event between Denisova and Papuans before it identifies one between MA-1 and Karitiana, even though the inferred level of shared genetic ancestry is much greater for MA-1 and Karitiana than it is for Denisova and Papuans. ... At 30 kya, population 2 receives 7.8% of its genetic material from population 1 and at 5 kya, population 4 receives 41% of its genetic material from population 3. The more ancient admixture event (7.8% mixture) represents a mixture between a pair of populations that are distant evolutionarily, whereas the more recent admixture event (41% mixture) represents a mixture between a pair of populations that are close evolutionarily.” In the Chinese Super-Grandfathers paper, the age estimation for the common ancestor of NO and P was 31-35kya. The recent event seems to be very important in size, and I really wonder what MA-1 like Siberian groups could ever have had such an impact in South America 5 kya.
According to Meyer et al 2012 (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3617501/) “Interestingly, we find that Denisovans share more alleles with the three populations from eastern Asia and South America (Dai, Han, and Karitiana) than with the two European populations (French and Sardinian). However, this does not appear to be due to Denisovan gene flow into the ancestors of present-day Asians, since the excess archaic material is more closely related to Neandertals than to Denisovans (Table S27). We estimate that the proportion of Neandertal ancestry in Europe is 24% lower than in eastern Asia and South America (95% C.I. 12–36%).”
“To increase the power to detect gene flow, we used a new approach, ‘enhanced’ D-statistics, which restricts the analysis to alleles that are not present in 35 African genomes and are thus more likely to come from archaic humans. This confirms that Denisovans share more alleles with Papuans than with mainland Eurasians. However, in contrast to a recent study proposing more allele sharing between Denisova and populations from southern China, such as the Dai, than with populations from northern China, such as the Han, we find less Denisovan allele sharing with the Dai than with the Han (although non-significantly so, Z = −0.9). Further analysis shows that if Denisovans contributed any DNA to the Dai, it represents less than 0.1% of their genomes today”.
ReplyDelete“Strikingly, Papuans share more alleles with the Denisovan genome on the autosomes than on the X chromosome (P=0.01 by a two-sided test). One possible explanation for this finding is that the gene flow into Papuan ancestors involved primarily Denisovan males”.
According to Reich et al. 2011 (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3188841/), “Aboriginal Australians, Near Oceanians, Polynesians, Fijians, east Indonesians, and Mamanwa (a "Negrito" group from the Philippines) have all inherited genetic material from Denisovans, but mainland East Asians, western Indonesians, Jehai (a Negrito group from Malaysia), and Onge (a Negrito group from the Andaman Islands) have not. These results indicate that Denisova gene flow occurred into the common ancestors of New Guineans, Australians, and Mamanwa but not into the ancestors of the Jehai and Onge and suggest that relatives of present-day East Asians were not in Southeast Asia when the Denisova gene flow occurred.”
According to Skoglund et al. 2011 paper (http://www.ncbi.nlm.nih.gov/pubmed/22042846/), Fig 1 E shows that there is again a higher frequency of Denisovan ancestry in New Guinea and Papua New Guinea and Northeast Australia (a link with mtDNA P?), but also in China and South America, excluding Mexico (areas with the highest mtDNA A and B frequencies), and also Japan and Amur seem less affected. We know that the most frequent haplogroups in New Guinea and Papua New Guinea are MNOPS derived M and S, and yDNA C2 is found mostly on the coast and not in the highlands as if it came to Guinea only after MNOPS. I suppose that yDNA D and C were not moving in the areas of the densest Denisovan concentrations as the Denisovan ancestry is absent in Onge, and Japan and Amur are less affected.
So is it possible that the whole MNOPS took the northern route and mixed with the Denisovans in a more northern area than usually thought (Central Asia, Altai) c. 30k years ago and the Denisovan-rich M and S ancestry was less diluted by later expansions on the remote Guinea Island? In Western Europe Denisovan ancestry became very diluted as most European haplogroups did not pass through Siberia/Denisovan areas. It is interesting that the highest Denisovan ancestry area in Europe seems to be in the areas with a lot of yDNA R1b and the lowest Denisovan ancestry area in the areas with a lot of yDNA I and J (Fig 1 E).
"The eastern branch, on the other hand, may have originated from deeper in Siberia and may have more closely resembled the Mal’Ta population".
ReplyDeleteI think that is probably correct. The presence of mt-DNA X may be the product of the diversion of the early people into the western North American cul de sac. As you say, 'The presence of mtDNA X is also a strong indication of real differences between the eastern and western Native American populations'.
"At least EDAR370A indicates the original Mogoloid population was the result of a unique hybridization event".
Granted that the geographic origin of the mutation given in the original paper may not be totally accurate I think we can assume that the population with it was responsible for spreading more genes than just that single one. I suspect that the gene actually introgressed into the modern human population through hybridization with an archaic population somewhere in the region, although no evidence for such a population has been found.
"The archaic genes may have diluted, but not the hybrid genes that became fixed in the East Asian type".
My guess is that the selection that gave rise to the homozygosity in the original population continued as the hybrid population moved further north and east.
"In the Chinese Super-Grandfathers paper, the age estimation for the common ancestor of NO and P was 31-35kya."
Stunningly close to the age given for the EDAR mutation's origin in modern humans, or introgression into them.
"perhaps the reason East Asians aren't particularly close to the boy from Malta is, their genetic profile comes from a later founding population that expanded with the Neolithic".
I'm sure that is the case, but is especially so for the movement south. The expansion north may have been somewhat earlier than the 'Neolithic'.
"relatives of present-day East Asians were not in Southeast Asia when the Denisova gene flow occurred".
Yes. That is what I have been trying (mostly unsuccessfully) to point out for years. Southeast Asians are largely the product of a Mongoloid phenotype that has moved south during the Neolithic. The earlier 'Papuan' phenotype from the region has been mostly replaced.
Sorry. Continued:
ReplyDelete"So is it possible that the whole MNOPS took the northern route and mixed with the Denisovans in a more northern area than usually thought (Central Asia, Altai) c. 30k years ago and the Denisovan-rich M and S ancestry was less diluted by later expansions on the remote Guinea Island?"
From my study of the migrations across Wallace's Line and into the Pacific I have come to the opposite conclusion. It was Y-DNA C and mt-DNA N who took the northern route, and it was eastward, and much earlier than 30k years ago. In greater Eurasia MNOPS-derived haplogroups managed originally to reach only as far north as Northern China/Inner Mongolia (in the form of NO) and back west and then north onto the Asian steppe (in the form of P, Q and R). From where Q and R expanded greatly.
"In Western Europe Denisovan ancestry became very diluted as most European haplogroups did not pass through Siberia/Denisovan areas".
That tends to indicate something other than what you envisaged above. The Denisova element wasn't 'diluted'. European MNOPS and mt-DNA R-derived groups' never contained it, except at the very beginning when they first left Sundaland. Only the ones that remained in Sundaland the longest picked it up from the newly-arrived Y-DNA C and mt-DNA N population.
"there is again a higher frequency of Denisovan ancestry in New Guinea and Papua New Guinea and Northeast Australia (a link with mtDNA P?)'
Actually a link with all mt-DNA Ns and Y-DNA C's in the region.
"We know that the most frequent haplogroups in New Guinea and Papua New Guinea are MNOPS derived M and S, and yDNA C2 is found mostly on the coast and not in the highlands as if it came to Guinea only after MNOPS".
Although, as you point out, New Guinea/Melanesian haplogroups mostly not Y-DNA C or mt-DNA N the MNOPS and mt-DNA M haplogroups there must have had time to mix with the Denisova-carrying population before themselves crossing Wallace's Line, thus carrying the Denisova element into New Guinea/Melanesia. There we go with admixture yet again.
Kristiina
ReplyDelete"Fig 1 E shows that there is again a higher frequency of Denisovan ancestry in New Guinea and Papua New Guinea and Northeast Australia (a link with mtDNA P?)".
I checked hg P and didn't find any excess in Denisovan/Neandertal matches. There are lineages such as U, X, L0 and some others that are much richer in those "archaic retentions," but not hg P.
"I guarantee I have had more experience with pratical genetics in all its forms than you have. "
The practical genetics of modern humans? You're a scary man, Terry.
German Dziebel writes:
ReplyDeleteAnd here's what an e-mail to me says:
"I think you'll be pleased to know that I do consider quite interesting and worthy of more thorough investigation indications of back-migration or complex bidirectional interaction between the New World and Eurasia, and believe that contemporary and ancient American genomic data may well play a useful role in disentangling archaic/modern interactions. I will also say that our group does have some reservations about the scenario Eske proposed.., given the potentially confounding factor of Amerindian-related North Asian gene flow into prehistoric Europe (as outlined in Nick Patterson's 2012 "Ancient Admixture in Human History"), which prehistoric West Eurasia > Americas input is by itself clearly insufficient to explain."
German, would you mind telling us whose e-mail message this is? I am 100% certain that it does not belong to David Reich.
Hamarfox, thanks for your responses.
ReplyDeleteNo problem. Having now had a decent look, the relationships are pretty intuitive. The Siberians that stream toward MA-1 represent the greater geographical spread. These same populations stream toward W. Eurasians in PCAs where MA-1 is absent, and in almost exactly the same order, suggesting an MA-1-like population is the overwhelming source of W. Eurasian ancestry in modern Siberians, or that MA-1 is the best available proxy for these influences (rather than modern Europeans or C/S Asians).
The Siberians streaming toward Amerindians belong to contact zones in NE Siberia and Greenland. These populations are as related to MA-1 as the populations streaming directly toward him, so they seem to be related to him by way of Amerindians. Consistently, these populations also have significant W. Eurasian ancestry (present in Dodecad k=3 but consumed by 'Amerindian' at k=4 in, for example Chuckchi), so it seems they derive this also by way of Amerindians. (I suspect they also received direct admixture from MA-1-like population, but I'm basing the above strictly on the PCA and a few other mentioned sources).
Terry and Rokus, did either of you know about this?
http://www.nature.com/news/mystery-humans-spiced-up-ancients-sex-lives-1.14196
There's not much detail, but since they have no archaelogical information (other than the fact that Denisova had some of it), they may well be basing claims of its Asian distribution on its specific relationship with Asian populations.
Terry and Rokus, did either of you know about this?
ReplyDeletehttp://www.nature.com/news/mystery-humans-spiced-up-ancients-sex-lives-1.14196
Other did as well. Hu et al. (2012): In the genealogy of each [non-African] archaic segment with Neanderthal, Denisovan, African and chimpanzee segments, 77~81% archaic segment coalesced first with Neanderthal, 4~8% coalesced first with Denisovan, and 14% coalesced first with neither
In Expanding Hybrids And The Rise Of Our Genetic Common Denominator I state it wouldn’t be farfetched to consider most of Hu’s 14% to be essentially archaic Asian and possibly indicative to the genetic contribution of up to four Asian archaic hominines. Actually, I deduced that shared African segments are indicative for a huge hybrid gene-flow from Europe into Africa and this was quite some time before it became clear that the Americas as well as Oceania apparently received an ever higher component of European signature. It seems sooner or later we'll have to redefine "AMH" and "European" to access the true nature of our genetic common denominator.
@ Onur
ReplyDeleteThat's what I felt comfortable sharing publicly and let's leave it at that. It was an answer to Dienekes's use of a press release to represent David Reich's perspective.
I have regular communications with geneticists working in the field both in the U.S. and in Russia. Every now and then I bring up their names and opinions. Recently, this kind of information got misused at one of the popular science forums (see my post on Anthropogenesis regarding Anthrogenica folks) and I don't want this to happen again.
Ultimately it doesn't matter what this or that geneticist thinks (no offense). It's up to broadly trained anthropologists to make conclusions. What matters is the data that geneticists uncover. They can follow the data or they can follow the consensus lore common in archaeological or genetic communities or they can look for a diplomatic middle ground. But the data for an out-of-America migration is available and it's pretty straightforward and it has found its advocates within and outside of academia. Let's just see which academic scholar will choose which path.
@hamarfox -
ReplyDeleteYeah, it seems like all the Siberians streaming towards Amerindians do belong to the contact zones you discuss, while those streaming towards MA-1 are those who are geographically distributed (although tending to be Western) ...
.. however the most MA-1 like sample is the Aleutian, which is right there about as close to North America as you can get, but a bit off the beaten track. Isolated relict?
Looking closer at the Aleutians and the PCA, it seems striking that the Amerindian populations appear intermediate between Aleutian and Karitiana. There's an Aleutian-Karitiana cline and a Aleutian-East Asian cline.
By contrast, there is a "no-mans land" between Karitiana and the Eastern Greenlanders, who look like the foreshortened pole of a direct East Asian-Karitiana admixture cline.
If we look at the ADMIXTURE Dienekes has previously posted up on his blog from the paper for the "Saqqaq" "paleo-eskimo" genome, from northwest Greenland 4000 years ago, this shows up without any European, present in the Aleutian sample in that same ADMIXTURE.
Today, judging by this paper's PCA, the Western Greenlanders incline away from the Eastern Greenlanders in a more Aleutian direction. They appear on this to a first approximation like Aleutian mixed East Greenlanders.
I can't draw any conclusions or tender any hypotheses from this at all...
That's what I felt comfortable sharing publicly and let's leave it at that. It was an answer to Dienekes's use of a press release to represent David Reich's perspective.
ReplyDeleteGerman, the way you publically shared it was confusing. I, and I am sure many others, initially thought that it belonged to David Reich from the way you presented it. Now I know that it does not belong to Prof. Reich, but that is because of my e-mail correspondence with Prof. Reich during the last couple of days. Now, after my e-mail correspondence with Prof. Reich, I am 100% sure that Dienekes' representation of Prof. Reich's views was fully accurate.
Here are the important parts of my e-mail correspondence with Prof. Reich:
Onur:
"I guess this question has been directed to you many times as of late, but, with your permission, I want to learn your, and your team's if possible, interpretation of the results of the new paper titled "Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans" (Raghavan et al. 2013) and of the conclusions of its authors, especially in the light of your papers Patterson et al. 2012 and Lipson et al. 2012, as apparently the conclusions of the authors of the Raghavan et al. 2013 paper directly contradict your and your team's conclusions in the Patterson et al. 2012 and Lipson et al. 2012 papers."
Prof. Reich's response:
"There is no contradiction; rather, the paper entirely complement each other.
My last two 2012 papers showed evidence of admixture into Europeans from an Ancient North Eurasian population related more closely to Native Americans than to any present-day East Asian population. We explicitly interpreted this as admixture into Europeans from a population that once extended from Europe to Siberia, and from there into the Americas before 15,000 years ago.
I think that Mal’ta is likely to be a representative of this population, and this would be entirely consistent with all three papers.
What is new in the Raghavan paper is the demonstration that Native Americans themselves are admixed from populations related to Mal’ta. However, this in no way contradicts the finding that there is also Mal’ta-like admixture into Europe. There is very strong evidence for admixture in both directions in my view."
And in another e-mail Prof. Reich wrote:
"I don’t have any disagreement with Eske Willerslev’s teams about the conclusions of their paper."
So it is clear as day that Prof. Reich completely agrees with Eske Willerslev's and his team's conclusions in the Raghavan et al. 2013 paper.
Of note, in a reply e-mail to my question about German Dziebel's some statements, Prof. Reich wrote this:
"I don’t know anyone named German Dziebel!"
No comment from me.:)
This comment has been removed by the author.
ReplyDelete@ Onur
ReplyDeleteI honestly don't understand why you've posted this. I posted what I posted and it was an actual piece of communication and it was relevant to what Dienekes had posted. I don't care if it was confusing to you.
"My last two 2012 papers showed evidence of admixture into Europeans from an Ancient North Eurasian population related more closely to Native Americans than to any present-day East Asian population."
This "ancient North Eurasian population" "related more closely to Native Americans than to any present-day East Asian population" is the same as Mal'ta and both are the same as Karitiana from the 2012 papers. Not Chukchees, not Chinese, but Karitiana. Raghavan et al. think that Amerindians are a mix of East Asians and West Eurasians. Reich postulates a third population that's neither but it admixed into West Eurasians.
There're two interpretations going on here, not one and the quote I gave you nails it on the head.
"So it is clear as day that Prof. Reich completely agrees with Eske Willerslev's and his team's conclusions in the Raghavan et al. 2013 paper."
It's clear as day, Onur, that your snooping around is sick.
""I don’t know anyone named German Dziebel!"
Well, now he does. We are all lifelong learners.
Onur, thanks very much for following that up.
ReplyDelete"The practical genetics of modern humans? You're a scary man, Terry".
So you're claiming that animal/plant genetics are somehow utterly different from human genetics? German: you are the scary one.
"The population genetics of modern humans is either part of anthropology (to which kinship studies and reflexive thinking are core) or it's nothing".
Really? Is that why they call it population'genetics'?
"it wouldn’t be farfetched to consider most of Hu’s 14% to be essentially archaic Asian and possibly indicative to the genetic contribution of up to four Asian archaic hominines".
Yes. I tend to think the number of 'archaic' populations that have contributed to modern human genes has been greatly under-estimated.
Terry quoted:
ReplyDelete"Furthermore, we estimate that 14 to 38% of Native American ancestry may originate through gene flow from this ancient population".
And then Terry commented:
So Native Americans can only be somewhere between 86 and 62% Mongoloid.
I say:
Mongoloids are, at least in part, a phenotypic class, and that phenotype could be acquired by people of any lineage that passed through the climates in which the Mongoloid phenotype as a selective advantage (namely, the very cold climates), as long as it stayed their long enough. People who acquired Amerind phenotype also passed through these climates and stayed for a time, but not as long as the Mongoloids, since they are not as cold-adapted. So both the peoples coming to Beringia from the west (with male Q and female A haplotypes) and those coming from the south (with female B, C and D) phenotypes passed through the cold climate of the tundra and taiga, and those who stayed their longest (such as Chukchians and Eskimo-Aleuts) become Monogoloids, whereas as those who passed out of the region sooner became Amerinds.
This comment has been removed by the author.
ReplyDelete@terryT
ReplyDelete"Really? Is that why they call it population'genetics'?"
It's a subfield within physical anthropology. There are other disciplines within the overall "four-field" anthropological approach with which anthropological genetics interacts. They all describe humanity in its biological and cultural manifestations. If you care to read a paper on the population genetics of human populations, you'll find references to archaeology, history, linguistics, ethnology, craniology. You won't find references to plant genetics. So, I don't know what "practical knowledge" of animal breeding or flower crossing can give you here.
I honestly don't understand why you've posted this. I posted what I posted and it was an actual piece of communication and it was relevant to what Dienekes had posted. I don't care if it was confusing to you.
ReplyDeleteWhat you posted was irrelevant to what Dienekes had posted since it did not come from David Reich. Using someone else's statements as representative of Prof. Reich's views is a dishonest behavior.
This "ancient North Eurasian population" "related more closely to Native Americans than to any present-day East Asian population" is the same as Mal'ta and both are the same as Karitiana from the 2012 papers. Not Chukchees, not Chinese, but Karitiana. Raghavan et al. think that Amerindians are a mix of East Asians and West Eurasians. Reich postulates a third population that's neither but it admixed into West Eurasians.
There're two interpretations going on here, not one and the quote I gave you nails it on the head.
There is only one interpretation. You have completely misunderstood Raghavan et al.'s interpretation. There is no difference between the intepretations of Raghavan et al. and Prof. Reich.
It's clear as day, Onur, that your snooping around is sick.
Looking at the heatedness of your response, now I am more confident than ever that I am on the right track.
German and Onur, what are your respective email addresses?
ReplyDeleteWhy did you ask that?
@Onur
ReplyDelete"Using someone else's statements as representative of Prof. Reich's views is a dishonest behavior."
I didn't say those were Reich's views. But I did mention his lab. Labs are referred to by the name of their chief researchers. But if you look at the 2012 papers that came out from this lab they are led by other researchers, with Reich being a contributing author.
"Looking at the heatedness of your response, now I am more confident than ever that I am on the right track."
A track to nowhere. I'm frustrated, Onur, for obvious reasons: whether Reich communicates different things to different people, or some of his team members don't share his views, or there's competition between his lab and Eske - you shouldn't be investigating it.
What I quoted here was from the right source and it was relevant to the discussion. And let's put an end to this conversation.
"There is only one interpretation. You have completely misunderstood Raghavan et al.'s interpretation. There is no difference between the intepretations of Raghavan et al. and Prof. Reich. "
I didn't misunderstand anything. Whether Raghavan and Reich are coherent in formulating their conclusions and the implications of what their labs have found is another matter.
Raghavan: "This suggests that populations related to contemporary western Eurasians had a more north-easterly distribution 24,000 years ago than commonly thought. Furthermore,we estimate that 14 to 38% of Native American ancestry
may originate through gene flow from this ancient population."
Reich: "What is new in the Raghavan paper is the demonstration that Native Americans themselves are admixed from populations related to Mal’ta. However, this in no way contradicts the finding that there is also Mal’ta-like admixture into Europe. There is very strong evidence for admixture in both directions in my view."
Raghavan is talking about a West Eurasian population that supposedly admixed into Amerindians, while Reich is talking about a Mal'ta population that admixed into West Eurasians and into Amerindians. He did smooth the corners here by saying that the papers that came out of his lab are complementary to Raghavan but the quote I brought up has a sharper edge:
"I will also say that our group does have some reservations about the scenario Eske proposed.., given the potentially confounding factor of Amerindian-related North Asian gene flow into prehistoric Europe (as outlined in Nick Patterson's 2012 "Ancient Admixture in Human History"), which prehistoric West Eurasia > Americas input is by itself clearly insufficient to explain."
My own perspective is even more radical of course. But also much more straightforward with the facts. Mal'ta is genetically a West Eurasian population that carries two kinds of Amerindian admixture (PINK and ORANGE). Neither modern Amerindians nor Mal'ta people have any East Asian admixture. So there's no "dual ancestry" of Amerindians, contra Raghavan, and there's no Mal'ta population distinct from West Eurasians and the same population can't admix into itself, contra Reich.
"Terry and Rokus, did either of you know about this?"
ReplyDeleteI hadn't seen the particular comment but I'm sure I remember something about it some time ago.
"I posted what I posted and it was an actual piece of communication"
Your 'logic' is extremely interesting, to say the least. Example:
"So there's no 'dual ancestry' of Amerindians, contra Raghavan, and there's no Mal'ta population distinct from West Eurasians and the same population can't admix into itself, contra Reich".
Both papers actually claim admixture, and neither claims the Mal'ta population is the only component in either population.
"Raghavan is talking about a West Eurasian population that supposedly admixed into Amerindians, while Reich is talking about a Mal'ta population that admixed into West Eurasians and into Amerindians".
The only difference between the two claims is that one doesn't consider Amerindians at all while the other does. In fact there is no difference between the two.
"Using someone else's statements as representative of Prof. Reich's views is a dishonest behavior".
Enough said.
"Mongoloids are, at least in part, a phenotypic class, and that phenotype could be acquired by people of any lineage that passed through the climates in which the Mongoloid phenotype as a selective advantage (namely, the very cold climates), as long as it stayed their long enough".
Not really so. You're claiming parallel evolution here but unless the people had genetic contact with people already 'Mongoloid' they would almost certainly develop a different phenotype that would achieve the same result. We see that in the difference between the European and East Asian responses to similar conditions. So a particular phenotype will only 'be acquired by people of any lineage that passed through' regions 'in which the Mongoloid phenotype' already existed.
I didn't say those were Reich's views. But I did mention his lab. Labs are referred to by the name of their chief researchers. But if you look at the 2012 papers that came out from this lab they are led by other researchers, with Reich being a contributing author.
ReplyDeleteI did not say that you had said that those were David Reich's statements. I said that you presented someone else's statements as if they were representative of Prof. Reich's views.
Prof. Reich was one of the leading authors of the 2012 papers of the Reich lab, and he is, as you say, the chief researcher of the Reich lab, so his views are certainly important.
A track to nowhere. I'm frustrated, Onur, for obvious reasons: whether Reich communicates different things to different people, or some of his team members don't share his views, or there's competition between his lab and Eske - you shouldn't be investigating it.
Prof. Reich said to me that he did not know anyone named German Dziebel, so he cannot be one of the researchers of the Reich lab who communicated with you. I have seen no contradiction in Prof. Reich's views. His support of Raghavan et al. 2013's conclusions are perfectly in line with his conclusions in the 2012 papers.
What I quoted here was from the right source and it was relevant to the discussion. And let's put an end to this conversation.
Those statements only represent the person who made them, not Prof. Reich or anyone else.
I didn't misunderstand anything. Whether Raghavan and Reich are coherent in formulating their conclusions and the implications of what their labs have found is another matter.
Raghavan: "This suggests that populations related to contemporary western Eurasians had a more north-easterly distribution 24,000 years ago than commonly thought. Furthermore,we estimate that 14 to 38% of Native American ancestry
may originate through gene flow from this ancient population."
Reich: "What is new in the Raghavan paper is the demonstration that Native Americans themselves are admixed from populations related to Mal’ta. However, this in no way contradicts the finding that there is also Mal’ta-like admixture into Europe. There is very strong evidence for admixture in both directions in my view."
Raghavan is talking about a West Eurasian population that supposedly admixed into Amerindians, while Reich is talking about a Mal'ta population that admixed into West Eurasians and into Amerindians. He did smooth the corners here by saying that the papers that came out of his lab are complementary to Raghavan but the quote I brought up has a sharper edge:
"I will also say that our group does have some reservations about the scenario Eske proposed.., given the potentially confounding factor of Amerindian-related North Asian gene flow into prehistoric Europe (as outlined in Nick Patterson's 2012 "Ancient Admixture in Human History"), which prehistoric West Eurasia > Americas input is by itself clearly insufficient to explain."
Raghavan et al. 2013 say "populations related to western Eurasians", not "populations like western Eurasians". They do not equate Western Eurasians with Mal'ta-like people. Unlike the 2012 papers, the focus of the Raghavan et al. 2013 paper is the ancestry of Native Americans, not the ancestry of Western Eurasians; that is why they do not deal with the ancestry of Western Eurasians that much.
Of course, more research is needed in this area, and I am sure that with the increase in the number of Palaeolithic-era modern human genomes studied, we will have much more robust knowledge and theories regarding the origin of modern human races and major groups.
@Onur
ReplyDelete"I said that you presented someone else's statements as if they were representative of Prof. Reich's views."
No I didn't. Dienekes pulled Reich's statement to the media to respond to my reference to the Reich Lab. I kept the original reference to the lab. You misread the conversation, Onur.
" his views are certainly important."
Sure. They're just not the only ones.
"His support of Raghavan et al. 2013's conclusions are perfectly in line with his conclusions in the 2012 papers."
No, they are not. He just wants them to be. Willerslev has a good rapport with the archaeological community who would like to see all the incoming genetic data not upset the current vision of the peopling of the America. The Reich lab is working on a series of ancient DNA from Eurasia, so we should wait and see where they net out.
"Those statements only represent the person who made them, not Prof. Reich or anyone else."
Not so. The quote refers to "our group."
"Raghavan et al. 2013 ... do not equate Western Eurasians with Mal'ta-like people."
Yes, they do because they write "populations related to contemporary western Eurasians had a more north-easterly distribution 24,000 years ago than commonly thought."
@terryT
"Both papers actually claim admixture, and neither claims the Mal'ta population is the only component in either population."
What two papers, Terry? There's Raghavan 2013. And then there are some statements from Reich, and then there are papers by Patterson 2012, Lipson 2013, and finally Reich 2012. Raghavan et al. don't even mention Patterson 2012 and Lipson 2013, which bring up Amerindian-like admixture in West Eurasians. (They discuss it in Suppl Mat only and the discussion is not very coherent.)
""Raghavan is talking about a West Eurasian population that supposedly admixed into Amerindians, while Reich is talking about a Mal'ta population that admixed into West Eurasians and into Amerindians".
"The only difference between the two claims is that one doesn't consider Amerindians at all while the other does. In fact there is no difference between the two."
To your pre-scientific ear only, Terry. Raghavan et al. claim that West Eurasians admixed into Amerindians after the latter had diverged from East Asians. West Asians are BLUE, East Asians are YELLOW. Neither color appears as a significant contribution to Amerindian population on the ADMIXTURE charts. Mal'ta's mtDNA is U and Y-DNA is R. Both clades have a West Eurasian distribution and neither of them appear in Amerindians.
When Reich is talking about Mal'ta, it makes more sense because he implies an extinct Siberian population that contributed a certain autosomal component to modern West Eurasians (referred to as "Amerindian-like" by Patterson et al. 2012 and Lipson et al. 2013) and to Amerindians. This component is ORANGE on Raghavan's ADMIXTURE chart and not BLUE.
Finally, since there are no modern populations in Siberia that would represent this component better than Amerindians and all Amerindians but Eskimo-Aleuts and Na-Dene are virtually fixed in this component (Reich et al. 2012 call it "First American"), it makes perfect sense to say that the Mal'ta population is part-Amerindian and that modern Eurasian populations are less Amerindian than Mal'ta.
Terry and Rokus, did either of you know about this?
ReplyDeletehttp://www.nature.com/news/mystery-humans-spiced-up-ancients-sex-lives-1.14196
I have previously brought heidelbergensis into this discussion. One possibility is that heidelbergensis mixed with E Asian erectus when they traveled east (perhaps from somewhat before to around 200,000 ya, when there are signatures of improved tools and much more prevalence of fire usage in China).
At the Denisova cave there where Neanderthals living roughly contemporaneously with this admixed population, so some "Denisovan" material might show Neanderthal admixture.
But when we talk about Densiovan admixture in humans, what we see might actually be the Asian erectus component. And when we talk about Neanderthal admixture into humans, what we see might in fact be (or partially be) the heidelbergensis component.
When AMH (Nubian culture) were in Arabia ~100,000ya, there was another stone technology just north of them. But Neanderthals had not spread that far east, yet, not for another ~40ky. So, admixture with heidelbergensis, locally there unadmixed with E Asian erectus, seems much more likely (they also likely looked less weird to AMHs than Neanderthals, were less drifted, and perhaps genetically closer to AMHs).
Finally, NE Asian erectus could have been cold-adapted compared to SE Asian erectus, which might explain a certain phenotype of (originally) NE Asian AMHs...
German, you checked hg P and did not find any excess in Denisovan/Neandertal matches. How about putative M29’Q or Australian M15?
ReplyDeleteWhen you said that lineages such as U, X, L0 and some others are much richer in those "archaic retentions, does it mean that mtDNA M includes less possible archaic elements? That would be significant, if there are indications that U and X were both moving early in Neanderthal areas.
NB: I noticed that numbering of M clades is confusing. In Genetic evidence for the colonization of Australia by Sheila van Holst Pellekaan 2011, Australian M haplogroups are M15, M14, M42 and a rare Q2. When I go to Wikipedia mtDNA M site, I see that M10 and M42 are grouped together under M10’42, and M74 has disappeared. Then, it is said that M14 and M15 are found in Tibet and M13 is a small clade found among Tibetans, Oirat Mongols in Xinjiang, Barghuts in Hulunbuir, and Yakuts and Dolgans in central Siberia. When I go to the Spanish M site, I see that M15 (o M13) is found in Australia and M14 in Australia and Saudi-Arabia. The Spanish site also says that M42 found in Australia and Austroasiatics and Dravidians in India, as well as M74 found in China, Indo-China and Philippines form a common M42’74 cluster.
ReplyDeleteIs anyone aware of changes involving relationship of Australian M clades with continental Asia?
No I didn't. Dienekes pulled Reich's statement to the media to respond to my reference to the Reich Lab. I kept the original reference to the lab. You misread the conversation, Onur.
ReplyDeleteIt is clear that that quote does not represent the Reich Lab as a whole, as David Reich, the principle invstigator of the Reich Lab, does not agree with it. You are attributing the personal views of someone, whether he/she is from the Reich Lab or not, to the whole Reich Lab. That is a dishonest behavior.
Sure. They're just not the only ones.
I am not attributing Prof. Reich's views to the whole Reich Lab.
No, they are not. He just wants them to be. Willerslev has a good rapport with the archaeological community who would like to see all the incoming genetic data not upset the current vision of the peopling of the America. The Reich lab is working on a series of ancient DNA from Eurasia, so we should wait and see where they net out.
Almost no one, including the researchers of the Reich Lab, takes your Out-of-America hypothesis seriously.
Not so. The quote refers to "our group."
But they are still statements of one person.
Yes, they do because they write "populations related to contemporary western Eurasians had a more north-easterly distribution 24,000 years ago than commonly thought."
"Related to" by no means means "equal to".
When Reich is talking about Mal'ta, it makes more sense because he implies an extinct Siberian population that contributed a certain autosomal component to modern West Eurasians (referred to as "Amerindian-like" by Patterson et al. 2012 and Lipson et al. 2013) and to Amerindians. This component is ORANGE on Raghavan's ADMIXTURE chart and not BLUE.
The "Amerindian-like" components of Patterson et al. 2012 and Lipson et al. 2012 in Western Eurasian populations are much bigger than the orange components of Western Eurasians in the ADMIXTURE analysis of Raghavan et al. 2013. That is because the orange component of Raghavan et al. 2013's ADMIXTURE analysis is a Native American component, whereas the "Amerindian-like" components of Patterson et al. 2012 and Lipson et al. 2012 are genetically somewhere between Native Americans and Western Eurasians. In the same way, the Mal'ta population is genetically somewhere between Western Eurasians and Native Americans. The Mal'ta population does not equal to the hypothetical Amerindian-like populations of the Patterson et al. 2012 and Lipson et al. 2012 papers, this causes some confusion. But there is no apparent error or contradiction in and between the interpretations of the results, as Prof. Reich says, the interpretations complement each other. Only more sampling and study of Palaeolithic genomes can fully clarify the degree and directions of the gene flows.
Here's a paper that supports the statement I posted earlier about significant differences between eastern and western branches on immigration into North America. Although this paper deals with mtDNA data, corresponding differences in Y-DNA groups may be inferred as well.
ReplyDeleteReconciling migration models to the Americas with the variation of North American native mitogenomes
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3761611/
A key quote from the Introduction:
"However, mitogenomes also show that Amerindians from northern North America, without any distinction between Na-Dene and non–Na-Dene, were heavily affected by an additional and distinctive Beringian genetic input. In conclusion, most mtDNA variation (along the double-continent) stems from the first wave from Beringia, which followed the Pacific coastal route. This was accompanied or followed by a second inland migratory event, marked by haplogroups X2a and C4c, which affected all Amerindian groups of Northern North America. Much later, the ancestral A2a carriers spread from Alaska, undertaking both a westward migration to Asia and an eastward expansion into the circumpolar regions of Canada. Thus, the first American founders left the greatest genetic mark but the original maternal makeup of North American Natives was subsequently reshaped by additional streams of gene flow and local population dynamics, making a three-wave view too simplistic."
"When I go to Wikipedia mtDNA M site, I see that M10 and M42 are grouped together under M10’42, and M74 has disappeared".
ReplyDeleteWiki is probably using a dated nomenclature. The Sheila van Holst Pellekaan 2011 nomenclature follows Phylotree.
"Is anyone aware of changes involving relationship of Australian M clades with continental Asia?"
The only connection noted so far is 'M42 found in Australia and Austroasiatics and Dravidians in India, as well as M74 found in China, Indo-China and Philippines form a common M42’74 cluster'.
"Australian M haplogroups are M15, M14, M42 and a rare Q2".
Correct. I'm sure you will find fig. 3 in this article interesting:
http://www.sciencedirect.com/science/article/pii/S1040618211002278
Unfortunately we still don't have data for SW or NE Australia but you will see that M14 and M15 are confined to Arnhem land with M15 also present in the Kimberly and central desert region. M42 (with relations in India and China) is, surprisingly, very prominent in the Riverine region.
"I have previously brought heidelbergensis into this discussion. One possibility is that heidelbergensis mixed with E Asian erectus when they traveled east"
I'm sure I remember that. And I think I remember commenting that the 'unknown' population may be the source of the EDAR mutation.
"Finally, NE Asian erectus could have been cold-adapted compared to SE Asian erectus, which might explain a certain phenotype of (originally) NE Asian AMHs..."
Fits perfectly. But that NE Asian erectus can not have had any Denisova element because that element is virtually basent in the region today.
"when we talk about Densiovan admixture in humans, what we see might actually be the Asian erectus component".
For reasons I mentioned above I have reason to doubt that to be so.
"To your pre-scientific ear only, Terry".
I believe I can safely ignore anything you say.
@German: it makes perfect sense to say that the Mal'ta population is part-Amerindian and that modern Eurasian populations are less Amerindian than Mal'ta.
ReplyDeleteNo, it doesn't make any sense at all. "Amerindian" didn't even exist when Mal'ta was alive - it would be like saying that Denisovans are part-Melanesian or Cro-Magnon are part-Basque.
Also, many modern East Asian populations would be "more Amerindian" than Mal'ta... perhaps you meant "European", not "Eurasian"?
I said:
ReplyDelete"Mongoloids are, at least in part, a phenotypic class, and that phenotype could be acquired by people of any lineage that passed through the climates in which the Mongoloid phenotype as a selective advantage (namely, the very cold climates), as long as it stayed their long enough".
Terry said:
"Not really so. You're claiming parallel evolution here but unless the people had genetic contact with people already 'Mongoloid' they would almost certainly develop a different phenotype that would achieve the same result. We see that in the difference between the European and East Asian responses to similar conditions. So a particular phenotype will only 'be acquired by people of any lineage that passed through' regions 'in which the Mongoloid phenotype' already existed."
I said:
I could believe in the developing of a different phenotype to achieve the same result if the lineages were very diverse (belonging to different species and probably different genera and familes), but amongst two lineage of Homo sapiens, whose racial variations are mostly not huge, it seems unlikely to me. I think that the main difference between European and East Asian phenotypes is due to the fact that the predominant East Asian phenotype originated mostly elsewhere, in a colder climate, colder than all but the northernmost and northeasternmost parts of Europe.
However, even if you are right in thinking that the Mongoloid features of later Mongoloids were acquired through admixture with earliest Mongoloids, it is still the case that only North Asia is cold enough for the earliest Mongoloids to acquire the Mongoloid phenotype, as I argued in the reply I just posted on your other thread, concerning MNOPS. Again, in brief: East Asia and even Mongolia cannot be the Mongoloid homeland, because people in equivalent climates to the west are not Mongoloid.
@Onur
ReplyDelete"It is clear that that quote does not represent the Reich Lab as a whole, as David Reich, the principle invstigator of the Reich Lab, does not agree with it. You are attributing the personal views of someone, whether he/she is from the Reich Lab or not, to the whole Reich Lab. That is a dishonest behavior."
There's nothing dishonest about my writing. No matter how hard you are trying to portray it in this way. The quote I gave clearly refers to "our group," which means it's not an opinion of a single person. Even it were, it would only illustrate my point that Reich's opinion is not the only one. Your behavior of snooping around, pitting one scholar against another, twisting reality and denying the veracity of my sources is clearly dishonest and inflammatory. You have no business being part of this conversation in the first place.
"The Mal'ta population does not equal to the hypothetical Amerindian-like populations of the Patterson et al. 2012 and Lipson et al. 2012 papers, this causes some confusion. But there is no apparent error or contradiction in and between the interpretations of the results, as Prof. Reich says, the interpretations complement each other."
I just showed you that they don't complement each other, but rather one is the evolution of the other toward an out-of-America angle. (I wish Reich was part of this conversation.) The Mal'ta population as a whole doesn't equal an Amerindian population but it carries 2 Amerindian components. From the ADMIXTURE graph we can see that the ORANGE component in Mal'ta is bigger than the ORANGE component in modern West Eurasians. Same for PINK. This is ABSOLUTELY what we would expect because Mal'ta is geographically closer to America than Western Eurasian.
@Tobus
""Amerindian" didn't even exist when Mal'ta was alive - it would be like saying that Denisovans are part-Melanesian or Cro-Magnon are part-Basque."
No. It's East Asians apparently didn't exist (at least not in Siberia from where they were supposed to enter America) when Mal'ta and Amerindians were around because neither Mal'ta, not Amerindian have the YELLOW component. The attestation of the Amerindian components in Ma'ta is proof of the Amerindian antiquity beyond the 12,000 YBP time frame suggested by the Clovis-first archaeological horizon.
We clearly need more ancient DNA samples to develop a genetic nomenclature that's not rooted in modern ethnic and racial names. But there's no logical contradiction in saying that Mal'ta has an Amerindian component. If we obtain DNA from a Paleoindian skull that will have an East Asian component (modal in Chinese) than we'll be able to say that Paleoindians were part Asians. But you may prefer to say that Chinese are Paleoindians because Chinese didn't exist 12,000 years ago. It'd be fair to say that Denisovans and Melanesians are related and that Melanesians are an older population than East Asians who, as generally considered, lack the Denisovan component. Similarly, Amerindians look older than East Asians because the East Asian component is absent in the oldest modern human remains in (North)East Asia.
@TerryT
"I believe I can safely ignore anything you say."
Good. I realized that the best way to fight "trolls" is to give them a piece of science. I hope poor Maju is reading this.
@kristina
ReplyDeleteI'm traveling through December 7. I will look up answers to your questions once I'm back. I was indeed surprised to see so many matches between Neanderthal / denisovan sequences and human hg U and X. But more work is needed to make current human mtdna phylogenies in sync with ancient hominid DNA data.
"I checked hg P and didn't find any excess in Denisovan/Neandertal matches. There are lineages such as U, X, L0 and some others that are much richer in those 'archaic retentions,' but not hg P".
ReplyDeleteKristiina. As Onur and Tobus show I would place as much reliance on that comment as I would on anything else German might say.
"Good. I realized that the best way to fight 'trolls' is to give them a piece of science. I hope poor Maju is reading this".
I'm pleased you brought up Maju. Like you he is absolutely committed to his particular belief and totally dismisses any research that contradicts this committment. How does the link Gary Moore recently provided fit your claim?
"Here's a paper that supports the statement I posted earlier about significant differences between eastern and western branches on immigration into North America".
Great paper, thanks. There has been a shortage of research that pinpoints various haplogroups within America. The only evidence at all here of any mt-DNA haplogroup having entered Siberia from America is:
"Our analyses reveal that the female effective size of these ancestral Paleo-Eskimo populations underwent a detectable bidirectional expansion temporally starting from approximately 4 ka (Fig. S2), covering all of the artic regions both in northern Canada and Greenland (on the American side) and in northeastern Siberia (on the Asian side), as a result of the back-migration process proposed by Tamm et al. (8). This expansion event is the same that led to the spread of: (i) mtDNA haplogroup D2a1, found in the northernmost Eskimos (and the single individual available from the extinct Paleo-Eskimo Saqqaq culture, dated to 3.4–4.5 ka; but hopefully other Paleo-Eskimo remains will be found and analyzed soon), Chukchi, Aleuts, Athapaskans, and possibly the Tlingit (21, 61); and (ii) Y-chromosome haplogroup Q1a*-MEH2, found in the Koryaks and Chukchi of Northeast Siberia, as well as in the ancient Paleo-Eskimo already mentioned above"
And I remember German being utterly opposed to my suggestion of some time back along the lines of:
"The estimated age of B2a and its localized geographic distribution indicate that the mutational motif of B2a most likely evolved in one of the (probably numerous) derived population groups that arose locally in North America along the trail of the coastal migration route. The early B2a carriers might have initially diffused along the northwestern Pacific coastal region, eventually resulting in a major in situ expansion in the (greater) American Southwest and northern Mexico"
@ Gregory76:
"even if you are right in thinking that the Mongoloid features of later Mongoloids were acquired through admixture with earliest Mongoloids, it is still the case that only North Asia is cold enough for the earliest Mongoloids to acquire the Mongoloid phenotype"
'Cold' alone is not sufficient to explain Mongoloid features. The phenotype also shows adaptation to a highly reflective environment, probably indicating high altitude as well as high latitude. Just such an environment exists within the region the EDAR370A mutation is claimed to have entered the modern human gene pool: bounded in the north by the Ordos Plateau, the west by eastern Tibet, the south by the Tsin Ling Shan and in the east by the western margin of the North China Plain.
"Again, in brief: East Asia and even Mongolia cannot be the Mongoloid homeland, because people in equivalent climates to the west are not Mongoloid".
But they are Mongoloid in all regions of reasonable altitude as far west as the Altai. As well as regions to the north of course.
There's nothing dishonest about my writing. No matter how hard you are trying to portray it in this way. The quote I gave clearly refers to "our group," which means it's not an opinion of a single person. Even it were, it would only illustrate my point that Reich's opinion is not the only one. Your behavior of snooping around, pitting one scholar against another, twisting reality and denying the veracity of my sources is clearly dishonest and inflammatory. You have no business being part of this conversation in the first place.
ReplyDeleteI just kindly asked you the source of your information, and you did not reveal. I did not criticize you for not revealing your source but for using your information in a misleading way. Also I never stated that Prof. Reich's opinion is the only one. I said that Prof. Reich's opinion is more or less the same as that of Raghavan et al. 2013. Opinions of some of the other researchers of the Reich Lab may be different, but they only represent themselves, just as Prof. Reich's opinion only represents Prof. Reich.
I see that you are now trying to retaliate by childishly trying to accuse me of the same misdeeds as the ones I accused you of and of more.
I just showed you that they don't complement each other, but rather one is the evolution of the other toward an out-of-America angle. (I wish Reich was part of this conversation.) The Mal'ta population as a whole doesn't equal an Amerindian population but it carries 2 Amerindian components. From the ADMIXTURE graph we can see that the ORANGE component in Mal'ta is bigger than the ORANGE component in modern West Eurasians. Same for PINK. This is ABSOLUTELY what we would expect because Mal'ta is geographically closer to America than Western Eurasian.
What you say by no means shows that the conclusions of the papers do not complement each other. The fact that the Mal'ta population has bigger Amerindian components than West Eurasians is something entirely expected and in no way contradicts Raghavan et al. 2013's conclusions.
@German:
ReplyDeleteNo. It's East Asians apparently didn't exist (at least not in
Siberia from where they were supposed to enter America) when Mal'ta and
Amerindians were around...
You seem very confused, there was no time "when Mal'ta and
Amerindians were around" - the Mal'ta speciment is from ~10kya before people entered the Americas, it didn't coexist with Amerindians.
"... because neither Mal'ta, not (sic) Amerindian have the YELLOW component"
The yellow component appears at K=2 *before* the orange at K=3 so "East Asian" certainly did exist before "American" did. I'm sure you know that each new colour as K increases on the ADMIXURE result represents a subset of the one of the previous colours, not a completely new unrelated lineage. Just like dark blue and green are both descend from the original light blue "West Eurasian" lineage, orange represents descendants of the earlier yellow "East Eurasian" lineage, not a completely independent ancestry. So it might seem confusing to you that Amerindians have no original yellow at K=11, but note that nearly all the colours they do have were split from yellow or yellow-descendants, indicating a common ancestor.
The attestation of the Amerindian components in Ma'ta is proof of the
Amerindian antiquity beyond the 12,000 YBP time frame suggested by the
Clovis-first archaeological horizon.
No it's not. It's an indication that there was a East Asian/Siberian population that was more similar to present day Amerindians than it was to present day East Asians. There is certainly no suggestion from the Mal'ta DNA that there were people in America any earlier than archaeological evidence suggests.
there's no logical contradiction in saying that Mal'ta has an Amerindian component
But that's not what you said.. you said "the Mal'ta population is part-Amerindian and that modern Eurasian populations are less Amerindian than Mal'ta". This is not correct because:
a) Amerindians didn't exist when the Mal'ta where around, so they can't be considered "part-Amerindian"... the best you can say is that Mal'ta were part-unknown-East-Eurasian-population-some-of-whose-descendents-eventually-populated-America.
b) modern East Asian populations are genetically closer to Amerindians than Mal'ta was, so in your terminology would be "more Amerindian" than Mal'ta, not less.
That new mtDNA study on Native American and in particular haplogroups A2a and B2a offers also an alternative origin for the American B2 haplogroup: ”an alternative explanation is that this B2 mtDNA entered the American continent through the ice-free corridor with the same population groups that introduced the C4c and X2a mitogenomes.
ReplyDeleteMost likely, the expansion of C4c and X2a in America occurred in the Great Plains region and our skyplot analysis suggests that the effective number of X2/C4c ancestral females began to increase from 8–10 ka (Fig. S2) right after the start of the stable, warm Holocene, which signed major postglacial expansions worldwide.”
The exciting thing about this is of course that both B and X are rare in Siberia.
According to Kumar et al. paper 2011, ”our analysis of a data set of 568 mitochondrial genomes from North, Central and South America, as well as Beringia and Siberia-Asia suggest that American founders diverged from their Siberian-Asian progenitors some time during LGM and expanded into America soon after the LGM peak (20-16 kya). Further, time between the Native American divergence from Siberian-Asians tothe expansion into America was shorter than the pre vious estimates. The phylogeography of haplogroup C1 suggest that this American founder haplogroup differentiated in Siberia-Asia. Although it is not clear for the haplogroup B2, haplogroups A2 and D1 might have differentiated soon after the Native American founder’s divergence. http://www.academia.edu/1021190/Large_scale_mitochondrial_sequencing_in_Mexican_Americans_suggests_a_reappraisal_of_Native_American_origins
When you look at Fig.4, you see that according to the models A and B, the average divergence time of Asian and American clades is set at 24 kya – 20 kya. It is just the time frame when Mal’ta boy like population existed in Baikal area. We all agree that Mal’ta boy seems to share ancestry with the Beringian populations that developed into Amerinds. The question is whether it was a result of admixture with the ancestors of Amerinds or a result of common ancestry.
On the basis of the admixture analysis, we see that Mal’ta boy shares two components with Native Americans, pink and orange. I now wonder if both these components existed already 24 kya? Is it possible that it is only a result of the structure of the modern populations? The Saqqaq individual, which is much more recent than Mal’ta boy, had three admixture components, of which none is the Amerindian component, but the main component could be the pink one (high in Koryaks), while the other two are Siberian and East Asian components. I think it is worth to note again that these two components seem to be more recent in Northeast Asia, developments taken place after the Ice Age (http://dienekes.blogspot.fi/2010/02/paleo-eskimo-whole-genome-sequenced.html)
Fits perfectly. But that NE Asian erectus can not have had any Denisova element because that element is virtually basent in the region today.
ReplyDeleteTerry,
From the Nature News article cited above:
"The Denisovan genome indicates that the population got around: Reich said at the meeting that as well as interbreeding with the ancestors of Oceanians, they also bred with Neanderthals and the ancestors of modern humans in China and other parts of East Asia. Most surprisingly, Reich said, the genomes indicate that Denisovans interbred with yet another extinct population of archaic humans that lived in Asia more than 30,000 years ago — one that is neither human nor Neanderthal."
I believe the interpretation I gave a while ago and summed up again above is the most parsimonious one that also fits archeological data.
a) Amerindians didn't exist when the Mal'ta where around
ReplyDeleteThis is an opinion, but what are your relevant arguments and how do you deal with counter evidence?
b) modern East Asian populations are genetically closer to Amerindians than Mal'ta was
If you had seen Figure 1 "PCA (PC1 versus PC2) of MA-1 and worldwide human populations" in the paper, you would have known this is utterly untrue.
I said:
ReplyDelete"even if you are right in thinking that the Mongoloid features of later Mongoloids were acquired through admixture with earliest Mongoloids, it is still the case that only North Asia is cold enough for the earliest Mongoloids to acquire the Mongoloid phenotype"
Terry said:
“'Cold' alone is not sufficient to explain Mongoloid features.”
I reply:
We can discuss whether it is sufficient (see below), but it is still necessary.
Terry continues:
“The phenotype also shows adaptation to a highly reflective environment, probably indicating high altitude as well as high latitude. Just such an environment exists within the region the EDAR370A mutation is claimed to have entered the modern human gene pool: bounded in the north by the Ordos Plateau, the west by eastern Tibet, the south by the Tsin Ling Shan and in the east by the western margin of the North China Plain.”
I reply:
If you are thinking of epicanthic folds and slanting eyes, that could be due to reflectance or it could be due to wind (or to wind plus snow, or wind plus sand, in the Kalahari). And if it is due to reflectance, it could be reflectance of snow (snow glare).
As to the region you describe, the same problem arises: regions to the west that are equally cold never evolved the Mongoloid phenotype.
I said:
"Again, in brief: East Asia and even Mongolia cannot be the Mongoloid homeland, because people in equivalent climates to the west are not Mongoloid".
Terry said:
“But they are Mongoloid in all regions of reasonable altitude as far west as the Altai. As well as regions to the north of course.”
I reply:
Yes, and west of the Altai? Mostly Caucasoid, at least at first.
And though, as you say elsewhere, Mongoloids replaced the Caucasoids in some of the western areas, it was replacement by invasion, not by evolution. So I say that such a climate could not be sufficient to produce the Mongoloid phenotype, or else the earliest people would be Mongoloids.
"they also bred with Neanderthals and the ancestors of modern humans in China and other parts of East Asia".
ReplyDeleteThanks. I was certainly under the impression that the element was absent through East Asia.
"Most surprisingly, Reich said, the genomes indicate that Denisovans interbred with yet another extinct population of archaic humans that lived in Asia more than 30,000 years ago — one that is neither human nor Neanderthal".
And I would guess that the population in question is the one in which the EDAR370A mutation existed.
"I believe the interpretation I gave a while ago and summed up again above is the most parsimonious one that also fits archeological data".
Do you mean this summary:
"This is consistent with the idea that the admixture went underway farther East, in extreme E Siberia and Beringia, with a brother group (Q, X) of Mal'ta boy, and not his population, directly. So, IMO (as I stated before) a proto-Mongoloid admixture for the first Americans, and then successive Mongoloid admixtures for Na Dene and Inuits".
I totally agree with that scenario. My only disagreement with you has been over your use of the term 'proto-Mongoloid'. To me it is quite possible that the 'extinct population of archaic humans that lived in Asia more than 30,000 years ago — one that is neither human nor Neanderthal' was probably the source for many of what we now know as Mongoloid features.
@Tobus
ReplyDeleteYou are the one who is confused. Archaeology tells us nothing about when America was peopled. Read my website about Toca de Tira Peia dated at 22 000 ya. The archaeological record in the Americas is growing in age just a bit slower than in other continents. The Amerindian components in Malta is just another evidence for that. The earliest Mongoloid skulls appear in the new world and not in Asia.
Regarding your reading of the Admixture results, the source of your confusion is that they colored South American Indians in yellow just like East Asians at k = 2 although they could have just as well colored Amerindians orange at k = 2. Yellow is just as much an Amerindian component as it is East Asian one in raghavan 2013. Go to my website and read the most recent post about Malta where I reproduce the cluster analysis from Rosenberg et al. 2002 in which Amerindians are there all the way down to k = 2.
What matters is that yellow doesn't appear in Amerindians at higher k levels. Neither does it appear in Malta. If orange is derived from yellow then it would make no sense for raghavan to propose the dual origin of Amerindians and Malta would have been derived from East Asians, too.
Another support point for this interpretation of what was presented in raghavan is that tiyanyuan mtdna is ancestral to all Asians but the closest living population to tiyanyuan is Karitiana.
@ onur
You called me dishonest. For no reason. Now you call me childish. For no reason. I didn't want to disclose the identity of my correspondent for the reasons I identified above. If people don't answer your question, it's not the reason for you to start snooping around. You are being utterly offensive.
@onur,
ReplyDelete"I don’t have any disagreement with Eske Willerslev’s teams about the conclusions of their paper."
"any present-day East Asian population"
If so, Reich is clearly going away from his prior paper's conclusions if I read the term present day correctly. Reich now by agreeing with Willerslev is disagreeing with himself.
Reich and collaborators: “The most striking finding is a clear signal of admixture into northern Europe, with one ancestral population related to present day Basques and Sardinians, and the other related to present day populations of northeast Asia and the Americas.”
"Yes, and west of the Altai? Mostly Caucasoid, at least at first. And though, as you say elsewhere, Mongoloids replaced the Caucasoids in some of the western areas, it was replacement by invasion, not by evolution".
ReplyDeleteWest of the Altai the altitude diminishes considerably. So the dividing line between Mongoloid and Caucasoid has become an altitudinal difference. Surely that is obvioulsy why 'regions to the west that are equally cold never evolved the Mongoloid phenotype'. And replacement by invasion is still replacement.
"So I say that such a climate could not be sufficient to produce the Mongoloid phenotype, or else the earliest people would be Mongoloids".
Correct. The fact that the earliest people west of Altai were not Mongoloid surely tells you that cold alone cannot have been responsible for the Mongoloid phenotype. Although the earliest people into northeast Eurasia were Caucasoid they came from west of the Altai, by 'invasion' if you like. They were presumably able to be first into the region because it was uninhabited at the time. The Mongoloid people had been unable to move as far north as the steppe dwellers from further west. However once the Mongoloid people had adapted the incoming technology they were in turn able to replace the Caucasoid people.
"If you are thinking of epicanthic folds and slanting eyes, that could be due to reflectance or it could be due to wind (or to wind plus snow, or wind plus sand, in the Kalahari). And if it is due to reflectance, it could be reflectance of snow (snow glare). As to the region you describe, the same problem arises: regions to the west that are equally cold never evolved the Mongoloid phenotype".
Yes. Regions to the west are much lower in altitude and do not have the same level of reflectance, either through semi-desert conditions or through snow. In northern Europe the reflective nature of the winter snow-covered environment was alleviated considerably by the presence of forest. So the conditions did not lead to anything like the Mongoloid phenotype.
You called me dishonest. For no reason. Now you call me childish. For no reason. I didn't want to disclose the identity of my correspondent for the reasons I identified above. If people don't answer your question, it's not the reason for you to start snooping around. You are being utterly offensive.
ReplyDeleteI did not call you dishonest and childish because of your concealing of the identity of your correspondent, they have nothing to do with that. I already explained in my previous posts why I called you dishonest and childish, so I won't repeat myself. Anyway, this discussion has gone on too long, so I am finishing it here.
Matt,
ReplyDelete.. however the most MA-1 like sample is the Aleutian, which is right there about as close to North America as you can get, but a bit off the beaten track. Isolated relict?
The Aleutians are from the same sample as population 160 here:
https://docs.google.com/spreadsheet/ccc?key=0ArJDEoCgzRKedGR2ZWRoQ0VaWTc0dlV1cHh4ZUNJRUE#gid=0
70% European. Without biographical information of the samples, it's really hard to interpret. It's all the weirder considering Raghavan rejected samples with recent admixture. Was this an oversight, or are Aleutians Mal'ta-like modern hold outs? One thing I noted was that the Aleuts have W. Eurasian components MA-1 doesn't seem to have, based on what analyses I've seen using the Dodecad calculator (on genetiker's blog). The ratio of W. Eurasian components (for example Southern to West Asian to Atlantic Baltic at k=12) better fits with admixture with a population more similar to modern Russians. Unfortunately, Aleuts don't seem to be included in any calculator for which pop. portraits are available, so I can't check to see whether the admixture is even across samples (i.e. old) or uneven. If it is recent, it doesn't explain why Aleuts aim at MA-1, but if it's old, that doesn't explain why Aleuts have components MA-1 doesn't.
Looking closer at the Aleutians and the PCA, it seems striking that the Amerindian populations appear intermediate between Aleutian and Karitiana.
My interpretation is that Amerindians stream toward an intermediate point between Mal'ta and East Asians, representing a spatial compromise between their two ancestries. This possibly intersects with some Siberian populations by chance?
The fact all Amerindians are fixed on a single point between MA-1 and East Asians suggests all Amerindian populations have roughly the same admixture proportions (and therefore the range of admixture in Amerindians offered by Raghavan may be the variation of the signal and not real variations in admixture).
If that's so, then the Karitianian pole of Amerindian variation may reflect an extreme of drift within America. If all Amerindians are essentially the same proportion of admixture between two ancestral populations, but differ in degrees of drift (and therefore 'unique Amerindian-ness'), I think that would explain the spatial distribution.
I'm not sure why some Siberians aim directly for Karitiana.
If we look at the ADMIXTURE Dienekes has previously posted up on his blog from the paper for the "Saqqaq" "paleo-eskimo" genome, from northwest Greenland 4000 years ago, this shows up without any European, present in the Aleutian sample in that same ADMIXTURE.
West Greenland is population 159 on the spreadsheet I linked to. Again, they are considerably more European than other Amerindian or Siberian groups, except Aleutians. The only not-completely-modern interaction with West Eurasians within the last 4000 years in Greenland would be with Scandinavians, who possibly just meet the requirements not to count as 'recent admixture'. Still, it's a stretch. The ratio of components did seem to match quite well with Norwegians on the k levels I checked.
@Rokus: This is an opinion, but what are your relevant arguments and how do you deal with counter evidence
ReplyDeleteIt's not just an opinion, it's the currect scientific consensus among experts in the field. There is no counter evidence (just fringe theories), which is probably why it's the consensus position.
If you had seen Figure 1 "PCA (PC1 versus PC2) of MA-1 and worldwide human populations" in the paper, you would have known this is utterly untrue.
Are you sure? Correct me if I'm wrong but that on that figure the X-axis (labelled PC1) represents the main component found, and it shows East Asians are closer to Amerindians than MA-1. It ranges from Europeans at one end (-0.04) to Karitiana at the other (+0.04), with the other Amerindians ranging from about +0.01-0.04. East Asians are at +0.1 with the Amerindians, Na Dene and Siberians while MA-1 is at -0.01 in between Oceanians and South Asians. This means that the main genetic variance in the group is between Europeans and Native Americans and that East Asians are closer to the Amerindians than MA-1 is.
The secondary component, shown on the "PC2" Y-axis, is the variance between East Asians and Native Americans and it's only in this secondary component that MA-1 is closer to Native Americans than East Asians are. This "closeness" only appears when the primary European-Ameridian component is removed. The 3rd and 4th components found (in Figure SI 9) show East Asians and Native Americans grouped very closely together a long way from MA-1. This means that East Asians are closer to Amerindians than MA-1 is and it's only when you ignore this difference and look that the variance between the two that MA-1 appears closer to Amerindians.
This is supported by the next plot (SI 10 B/C/D) where Amerindians are shifted significantly further towards the Han (0.02-0.08) than they are towards MA-1 (0-0.025). Further corroboration comes from the other analysis methods which also show East Asians are closer - the f3 Shared Drift plot (Fig SI 26) shows higher f3 results for Amerindians/Han and East Asians/Karitiana than MA-1 with either Han or Karitiana, and the ADMIXTURE anaylsis (Fig SI 7) which shows Amerindians are almost completely comprised of the yellow-derived (East Eurasian) colours while MA-1 is mostly blue-derived (West Eurasia) with only a small amount of yellow-derived components.
Contrary to what you claim, my statement that modern East Asian populations are genetically closer to Amerindians than Mal'ta appears to be completely correct.
AP,
ReplyDeleteIf so, Reich is clearly going away from his prior paper's conclusions if I read the term present day correctly. Reich now by agreeing with Willerslev is disagreeing with himself.
I agree. By not considering Amerindians and NE Siberians to be admixed populations, Reich & co. inferred that the related variation between Amerindians and W. Eurasians was of the same basic origin as the majority ancestry of Amerindians (i.e. East Asian). This led to incorrect quantifications of East Asian admixture in West Eurasia. Reich seems now to acknowledge this error, but his current position is unclear.
Willerslev definitely seems to favour the interpretation that Mal'ta was West Eurasian, and not simply because his clan's descendants ended up mostly to the West. There's some talk in the main paper to this end (though not strictly equivalent to my following point), but even in the supplementary data, we see that some W. Eurasians (Mozabite, Palestinians etc.) have minimal shared drift with Mal'ta, yet those same populations are much more related to Karitiania than to Han. This makes the most sense to me if we consider Mal'ta 'putting a word in' for SW Asians, not through subsequent geneflow between ancient Siberia and SW Asia, but by being related to SW Asians on a more fundamental level; i.e. through both being essentially West Eurasian. Of course, MA-1s maternal and paternal haplogroups were likely stronger in SW Asia and N. Africa before recent geneflow from non-West Eurasian populations.
Terry, here's some more EDAR percentages you might not know about:
http://alfred.med.yale.edu/alfred/mvograph.asp?siteuid=SI663326A
Much bigger samples and a wider range of populations. Outside of East Asia (and the Americas) it seems coextensive and almost perfectly proportionate to ADMIXTURE estimates of the East Asian component, even down to the ~2% in Hungarians.
@German
ReplyDeleteSo "Archaeology tells us nothing about when America was peopled" but "the Amerindian components in Malta is just another evidence" of "the archaeological record in the Americas ... growing in age". Archaeology is useless and DNA from ancient Siberia improves the archaeology record in America? You're not really making any sense - you are supposed to be explaining how Mal'ta can be considered "part-Amerindian" when nobody was living in America until 10,000 years later... even if Toca de Tira Peia is genuine evidence of human habitation (which is being debated), it still puts the earliest Americans more recent than the Mal'ta site and fails to prove your point. Moreover the maximum estimated Amerindian Y-haplogroup coalescence is only 17kya so a 22kya population in America isn't going to be "Amerindian" as we currently understand the term, and is unlikely to be what we are seeing in the Mal'ta DNA anyway. Do you have any actual real evidence that Amerindians existed before 24kya, or is this just a pet theory of yours built from speculation upon speculation?
I reproduce the cluster analysis from Rosenberg et al. 2002 in which Amerindians are there all the way down to k = 2.
That analysis differs greatly from the one is this paper - it doesn't discern a European component until K=4 for instance. Comparing them, your one is from 2002 and uses 377 SNPs, this new paper is 10 years more mature and uses over 300,000 SNPs. It's fairly moot though, whether they were closer to modern East Asians or Amerindians, there were definitely people in East Asia at the time Mal'ta was alive, and you've provided no evidence the same can be said for America.
the closest living population to tiyanyuan is Karitiana
I assume this statement is based on Table 1 of Hu et. al 2013. You no doubt know that this is from a single partially reconstructed chromosome and not a genome wide analysis. While it's certainly useful as a pointer in the right direction it's not necessarily 100% accurate - for instance it says that Sardinians and French are closer to Dai, Han and Karitiana than any of those groups are to Tianyuan, and that both Europeans are closer to Papuans than to Tianyuan... both of which are unexpected. At any rate Karitiana being slightly closer to Tianyuan on that table doesn't prove (or even suggest) that there were people in America before the time of the Mal'ta specimen.
"The phenotype also shows adaptation to a highly reflective environment" what do snow and a ocean have in common ? how about shiny surfaces and and varying degrees of cold winds ? and probably varying degrees of lack of regular amounts of fat or oils depending on where they are confined too .
ReplyDeleteAP,
ReplyDeleteProf. Reich's 2012 papers were focused on the ancestry of West Eurasians, not that of Native Americans, plus there was no Mal'ta or Afontova Gora sample yet. Prof. Reich is correct in saying that the 2012 papers and the Raghavan et al. 2013 paper do not contradict each other and instead complement each other. I guess Prof. Reich and his team will make further studies on West Eurasian and Native American genetics in light of the Raghavan et al. 2013's findings. With more studies and sampling, the genetic picture will become more and more clear.
"Outside of East Asia (and the Americas) it seems coextensive and almost perfectly proportionate to ADMIXTURE estimates of the East Asian component, even down to the ~2% in Hungarians".
ReplyDeleteThat makes complete sense, and indicates the mutation spread with the expansion of an East Asian population. The only possibility I see for the southern element is the Neolithic expansion, and the only candidate haplogroups I can see are the Y-DNA O haplogroups with some minor mt-DNA haplogroups. And, of course, this paper shows its presence in the Americas is probably also connected with the presence of a non-Mal'ta East Asian element.
"'The phenotype also shows adaptation to a highly reflective environment' what do snow and a ocean have in common ? how about shiny surfaces and and varying degrees of cold winds ? and probably varying degrees of lack of regular amounts of fat or oils depending on where they are confined too".
There is no shortage of such environmental factors in Inner Mongolia and nearby. Besides which it is extremely doubtful that the Mongoloid phenotype developed any further north than Inner Mongolia. There is absolutely no evidence for any movement into SE Asia from anywhere north of Sichuan/Shanxi, whereas there is considerable evidence for southward movement from this latter region.
"Prof. Reich's 2012 papers were focused on the ancestry of West Eurasians, not that of Native Americans, plus there was no Mal'ta or Afontova Gora sample yet. Prof. Reich is correct in saying that the 2012 papers and the Raghavan et al. 2013 paper do not contradict each other and instead complement each other".
Absolutely. German is getting confused with his comments comparing the papers (deliberately?).
@Tobus
ReplyDelete"You're not really making any sense."
No, you are the one who makes no sense. Your appeal to past authority is useless in the light of the new data from Tianyuan, Mal'ta, Monte Verde, Toca de Tira Peia, Buttermilk Creek and other ancient sites. Your "consensus" is nothing compared to the power of facts. I'm not supposed to provide you with evidence that Mal'ta is part Amerindian. Raghavan et al. have already provided this evidence. You prefer to deny it - that's all.
In fact, you are the one who's supposed to provide a list of archaeological sites in Siberia with concrete, specific and unique archaeological signatures shared with the most ancient American sites indicating that the Siberian sites represent a proto-Amerindian population. These sites are supposed to explain the technnological and behavioral aspects of the most ancient New World sites including Clovis, Monte Verde, Toca de Tira Peia and others. They should also form an unbroken chain from a Siberian source through Alaska all the way to South America.
You haven't provided any such evidence. Simply because it doesn't exist. Your consensus builders have forgotten that they need to beef up their ideas about "the peopling of the Americas" with some hard facts.
" Do you have any actual real evidence that Amerindians existed before 24kya..."
Between the extent of linguistic diversity in the Americas (140 language stocks which is 2/3 of world linguistic diversity not counting PNG) and a number of archaeological sites in the Americas (who cares if they are debatable - everything in human origins is or should be debatable), yes, we do have more than enough evidence that Amerindians existed at 24,000 years. Toca de Tira Peia is 22000 YBP... in Brazil. It takes time to walk from Alberta to Brazil, you know...
Do you have any hard evidence for East Asians prior to 10,000 years? Any skull with Mongoloid features or something? All the earliest "Mongoloid" skulls are in America.
Such marquee phenotypical features as shovel-shaped incisors or EDAR are more prominent in Amerindians than in East Asians suggesting that they originated in the New World. At the same time such East Asian traits as blood group B, Y-DNA hgs NO and such phenotypical traits as epicanthus are NOT found in the Americas suggesting that East Asians were not the source population for Amerindians.
"That analysis differs greatly from the one is this paper - it doesn't discern a European component until K=4 ..."
These are all excuses: you can clearly see that the YELLOW component in Raghavan is just as prominent in South Amerindians a in Han, so it can just be ORANGE. Mal'ta at 24,000 YBP has ORANGE but doesn't have YELLOW, although it's geographically in East Asia. This is just a plain fact that you again choose to deny. Amerindians appear at K = 2 on all other STRUCTURE and ADMIXTURE plots published to date. Rosenberg was just the first one. It was fundamentally right in the way it portrayed worldwide genetic variation. The European component came later, but it's irrelevant for the present discussion.
"You no doubt know that this is from a single partially reconstructed chromosome and not a genome wide analysis."
Another excuse. The results are meaningful and until they are overturned by another analysis, they hold. Tianyuan mtDNA is hg B, which is widely attested in the Americas but is nearly missing in living Siberians. It's a perfect fit with the Mal'ta story.
"my statement that modern East Asian populations are genetically closer to Amerindians than Mal'ta appears to be completely correct."
Great. This only shows that East Asians are likely derived from the New World. The YELLOW component first appears in the New World among Na-Dene and Eskimo-Aleuts - these must be the remnants of the proto-East Asian population. This is consistent with craniology, linguistics and ancient DNA studies.
"And, indeed, both these haplogroups [R and U] are absent in Native Americans..."
ReplyDeleteI don't know that we know that, given that it seems to be standard practice to reject NA R Haplotype samples on the assumption that they come from late European admixture.
The fact all Amerindians are fixed on a single point between MA-1 and East Asians suggests all Amerindian populations have roughly the same admixture proportions (and therefore the range of admixture in Amerindians offered by Raghavan may be the variation of the signal and not real variations in admixture).
ReplyDeleteI agree, with some slight variation for more recent Na Dene, and excluding Inuit entirely. Of course, this is a strong argument supporting the previously found result that the First Americans spent a lot of time homogenizing in Beringia before even entering the Americas. Also, as I stated above, the West Eurasian admixture is clearly higher than estimated, here, because MA-1 is just a proxy for the true (Q, X) population, likely as much as 5 to 10ky removed from that one.
@German.
ReplyDeleteI said:my statement that modern East Asian populations are genetically closer to Amerindians than Mal'ta appears to be completely correct.
You replied: "Great"
Looks like we're getting somewhere, I take it you are ready to accept that the 2nd part of your original statement, "that modern Eurasian populations are less Amerindian than Mal'ta" is incorrect - East Asians are in fact closer to Amerindians than Mal'ta is.
I'm not supposed to provide you with evidence that Mal'ta is part Amerindian. Raghavan et al. have already provided this evidence
Excellent, now we're really getting somewhere. Raghavan et al. say "Furthermore, we estimate that 14 to 38% of Native American ancestry may originate through gene flow from this ancient population. This is likely to have occurred after the divergence of Native American ancestors from east Asian ancestors, but before the diversification of Native American populations in the New World"... this is confirmed by the D-statistic analysis described in SI 14.6 in the supplementary material which I suggest you read. If Raghavan is your evidence, then if anything Amerindians are "part-Mal'ta", not the other way round. The first part of your original statement: it makes perfect sense to say that the Mal'ta population is part-Amerindian is directly contradicted by the source you claim provides the evidence for it.
Between the extent of linguistic diversity in the Americas (140 language stocks which is 2/3 of world linguistic diversity not counting PNG) and a number of archaeological sites in the Americas (who cares if they are debatable - everything in human origins is or should be debatable), yes, we do have more than enough evidence that Amerindians existed at 24,000 years. Toca de Tira Peia is 22000 YBP... in Brazil. It takes time to walk from Alberta to Brazil, you know...
None of the sites you claim as evidence are pre-Mal'ta and the only one that comes close has yet to be confirmed and is lacking any human remains. As I said before (but you ignored) it also pre-dates the divergence of Amerindian haplogroups, so if even if it is genuine, it's probably not "Amerindian" in terms of this disussion anyway. As I thought, your theory that there were people in America before Mal'ta is based on speculation, not direct evidence.
On the other hand there are lots of confirmed archaeological sites that prove modern humans have occupied East Asian continously for at least the last 40,000 years, and both Y-DNA and mtDNA haplogroups support Amerindians being derived from these East Asian ancestors. So you'll have to excuse me if go with the facts instead of your speculation.
you can clearly see that the YELLOW component in Raghavan is just as prominent in South Amerindians a in Han, so it can just be ORANGE
I think I understand what you are saying - East Asians and Amerindians share the same component at K=3, yes? ... implying a common ancestor after the divergence of Europeans and Africans.
Mal'ta at 24,000 YBP has ORANGE but doesn't have YELLOW, although it's geographically in East Asia. This is just a plain fact that you again choose to deny.
I'm pretty sure I never denied that fact, just your speculation that this somehow means there were people in America at 24kya. In fact it implies the exact opposite - the ancestors of present-day Amerindians were in East Asia 24kya.
After reading your arguments I suspect part of your problem is that you assume the "East Asian" ancestry of Amerindians necessitates a strong affinity with present-day East Asians. It doesn't - one thing Mal'ta shows us is that the East Asian ancestors of Amerindians already had significant divergence from the East Asian ancestors of present-day East Asians before 24kya. There's no reason to expect Han (for example) and Amerindians to share 100% of their phenotype.
Is an admixture analysis of Tianyuan individual available? It should be instructive to compare Tianyuan and Mal'ta individuals, as they are both ancient and their results should not be biased by the structure of modern populations. I suppose that there is no admixture analysis of Kostenki individual. I hope that we will get soon admixture analyses of ancient specimens.
ReplyDeleteAfontova Gora belongs to haplogroup Q1a1 y-DNA and a unresolved branch of MACRO HG R MTDNA according to y-full.com
ReplyDeletethe R1a1 was apparently contamination, or a newer source.
http://yfull.com/full-genomes/
search for AF2.
http://snag.gy/S1RSQ.jpg is saved in case the data gets deleted.
@Tobus
ReplyDeleteI originally meant that Malta has more of the Amerindian component than west Eurasians.
I'm traveling now and don't have the paper and the suppl mat in front of me, so we will table our discussion about whether Amerindians are closer to East Asians or to Malta. ADMIXTURE shows special proximity between NaDene and Eskimo Aleuts and East Asians. Out of all the populations Malta is closer to Amerindians followed by northern west Eurasians.
Regarding Raghavans claims that their data shows gene flow from Malta to Amerindians and not the other way around, it's just a stereotype of thinking. It's plain obvious that Malta are part Amerindian, while Amerindians don't carry neither the mtdna, nor the y DNA nor the Main west eurasian component of Malta. Raghavan spends two pages trying to explain away this obvious fact and make it look like Amerindians are part Malta. One of their arguments is that the Amerindian component is found all over west Eurasia. But from an out of America perspective, this is precisely what you would expect. They also reference the fact that Amerindians are closer to East Asians, while Malta is further apart from the latter, hence gene flow must have gone from Malta to Amerindians and not the other way around. But they don't consider the possibility that East Asians themselves are derived from Amerindians and this is precisely what we can conclude from the intermediary position of Amerindians between East Asians and west Eurasians. A smaller contribution of Amerindian genes to Malta vs. East Asians or the origin of both from a subdivided ancestral pool in North America would result in precisely this kind of pattern.
You are behind times in looking for "confirmation" of archaeological evidence for the antiquity of man in the Americas. Archaeological record is by nature a fragmentary and ever changing one. And the sites around the world don't get confirmed by a supreme commission. Archaeological sites in America accumulate at slower pace than those outside of the Americas but this has nothing to do with the presence or absence of humans there. What's important is that we have evidence for Amerindians and not East Asians in Siberia at 24000 Ybp. If we get ADMIXTURE results from Paleoindian DNA that shows them to be 80% East Asians and 20% west Eurasians then you'd have a case for a dual origin of Amerindians. But we don't. What we have is evidence that Amerindians admixed into a west Eurasian population at a time when East Asians apparently didn't exist.
Your postulation of an imaginary non-East Asian but proto Amerindian population in East Asia is awkward because we have a real Amerindian population to work with. And it did have the feet to cross the Bering strait into Asia. There is no population in Asia that has the Amerindian component expressed stronger than in Amerindians. Just get rid of your anti-Amerindian bias and we will be all set.
@Kristiina
ReplyDeleteNo we don't have ADMIXTURE for Tianyuan.
@ Daniel
This will change things quite a bit. I was worried about contamination myself. Eske originally presented AG as having a much higher risk of contamination than Malta.
"one thing Mal'ta shows us is that the East Asian ancestors of Amerindians already had significant divergence from the East Asian ancestors of present-day East Asians before 24kya."
ReplyDeleteI think it actually shows the two populations basically had separate origins from soon after any OoA. Probably separate routes around the Himalaya/Tibetan Plateau.
"Regarding Raghavans claims that their data shows gene flow from Malta to Amerindians and not the other way around, it's just a stereotype of thinking. It's plain obvious that Malta are part Amerindian"
No. It's the other way round: Americans are part Mal'ta. That's why the researchers claim 'gene flow from Malta to Amerindians and not the other way around'. Under the scenario you propose Amerindians would have entered Asia and, remarkably, split into a Mal'ta and and East Asian complonent with no overlap between the two. Surely it is far more likely that Americans are a simple hybrid between a Mal'ta-type people and an East Asian people, with a greater contribution from the East Asian element. Mal'ta appears 'part Amerindian' because a similar population contributed to the American hybrid population.
"Amerindians don't carry neither the mtdna, nor the y DNA nor the Main west eurasian component of Malta".
Quite. Yet if their deeper origin was American that fact would be very difficult to explain away.
"Tianyuan mtDNA is hg B, which is widely attested in the Americas but is nearly missing in living Siberians. It's a perfect fit with the Mal'ta story".
B 'is widely attested in the Americas' but is also widely attested through South China, Southeast Asia and the Pacific islands. American B is just one tiny branch of that hugely widespread haplogroup. American B is part of B4b, which is part of B4b'd'e'j, which is the only branch of B, of any sort, found outside South China/Southeast Asia. And yet you're claiming it originated in America. The fact B is 'nearly missing in living Siberians' suggests it did not at any time move through Beringia. Some B haplogroups are present in Japan so it is reasonable to assume B enetered America (or left it!) via the coast. This fits the proposal made in a link Kristiina provided elsewhere a few days ago.
This comment has been removed by the author.
ReplyDeleteThis comment has been removed by the author.
ReplyDeleteThis comment has been removed by the author.
ReplyDeleteThis comment has been removed by the author.
ReplyDeleteApologies. It was Gary Moore who provided the relevant link:
ReplyDeletehttp://www.ncbi.nlm.nih.gov/pmc/articles/PMC3761611/
Just to remind German:
And I remember German being utterly opposed to my suggestion of some time back along the lines of:
"The estimated age of B2a and its localized geographic distribution indicate that the mutational motif of B2a most likely evolved in one of the (probably numerous) derived population groups that arose locally in North America along the trail of the coastal migration route. The early B2a carriers might have initially diffused along the northwestern Pacific coastal region, eventually resulting in a major in situ expansion in the (greater) American Southwest and northern Mexico"
Note: 'population groups that arose locally in North America along the trail of the coastal migration route'.
@German:
ReplyDeleteI originally meant that Malta has more of the Amerindian component than west Eurasians.
I accept your retraction.
Regarding Raghavans claims that their data shows gene flow from Malta to Amerindians and not the other way around, it's just a stereotype of thinking
You're the one who claimed Raghavan as supporting evidence. I take it you withdraw that claim?
It's plain obvious that Malta are part Amerindian
Using the term "Amerindian" like this when talking about the time of Mal'ta is misleading - the "Amerindian" component at that time was was not an American population either geographically or genetically (nor was it Indian, but we can blame that misnomer on Columbus). It's called "Amerindian" because present-day Amerindians are the only surviving descendants of it, not because it originated or somehow belongs to America. Mal'ta can only be considered "part-Amerindian" if the meaning of "Amerindian" is changed to something like "ancient Siberian".
But they don't consider the possibility that East Asians themselves are derived from Amerindians
That's because it's not a possibility. Amerindians and present-day East Asians share a common Tianyuan-like ancestor that lived in Asia (not America) 40+kya. Mal'ta tells us they diverged no later than 24kya in Asia (again, not America) and ADMIXTURE shows negligible admixture since then. American Y and mt haplogroups indicate a common ancestor at around ~15kya, so without a time machine it's just not possible that Amerindians are the ancestors of East Asians... unless you want to redefine "Amerindian" to mean something very different.
Archaeological sites in America accumulate at slower pace than those outside of the Americas but this has nothing to do with the presence or absence of humans there
... unless there's nothing there to find of course, then it would be entirely to do with the presence or absence of humans there.
What's important is that we have evidence for Amerindians and not East Asians in Siberia at 24000 Ybp
.. and yet we have zero evidence for Amerindians in America at 24000 Ybp... you know what that suggests?
Your postulation of an imaginary non-East Asian but proto Amerindian population in East Asia...
"Imaginary"? (Psst: "we have evidence for Amerindians...in Siberia at 24000 Ybp")
... is awkward because we have a real Amerindian population to work with
Not at 24kya we don't.
Just get rid of your anti-Amerindian bias and we will be all set.
That's an exremely strange way to put it... should I assume from this that you harbour an anti-Asian bias?
Eastern View,
ReplyDeleteJust some straight-forward facts out of pitying yet another so far unwarranted appropriation of Native American heritage:
1. The "raw data", cluster analysis, shows Mal'ta to be an admixed individual not forming a cluster of its own. It shows no admixture in pure Amerindians.
Some ADMIXTURE runs show Amerindians to be unmixed at a deep level, while others show them to be admixed. Here are two calculators that show Amerindians to be admixtures of Asians and Europeans at k=3:
https://docs.google.com/spreadsheet/ccc?key=0AuW3R0Ys-P4HdHdGXzdLR0prSk1UUWZjNGl5cUEtWlE#gid=1
and:
https://docs.google.com/spreadsheet/ccc?key=0ArJDEoCgzRKedGR2ZWRoQ0VaWTc0dlV1cHh4ZUNJRUE#gid=0
A West Eurasian admixture is strictly Raghavan's own interpretation of the data, which is not the most straight-forward interpretation.
It is. In fact, I and several other people predicted an admixed origin of Amerindians based on the available evidence before Raghavan's paper.
The tree Raghavan generates is based on a premise
a. that Mari, Europeans, Indians, SW Asians form a population X. This is problematic because there never was such a population. The Uralic Mari are 30% Siberian in cluster analysis
Which is why migration edges are required to (accurately) explain population differences.
The resulting interpretation creates a huge paradox:
a. Uralic, Siberian, and groups known to have undergone high paternal European admixture (Aleut) are closest to Mal'ta.
By this logic, Uralic peoples, other Siberians, and Aleuts are the purest populations while Europeans and Amerindians are admixed. This is like saying the Uighur are the purest population and populations to its east and west are admixed.
They're closest to Mal'ta because they received direct admixture from a Mal'ta-like population. There are two ways a population can share drift with Mal'ta: have direct admixture with his clan (or a similar one), as is true of Amerindians, NE Siberians, and Siberians in general (in that order); or be related on a deeper level with MA-1, regardless of admixture, as with N Europeans, S. Europeans and W. Asians, in that order.
Kristiina,
ReplyDeleteIs an admixture analysis of Tianyuan individual available?
It's in the most recent post here:
http://genetiker.wordpress.com/
I'm wary of ideological bloggers, but there's some interesting analyses there that I've not seen elsewhere.
There are a few things I always consider when loooking at ancient DNA ADMIXTURE results: Ancient samples often appear as admixtures of populations that they in fact contributed genes to (as with the 'Amerindian' in MA-1 and Afontova Gora further down the page), while other times the admixture is likely real (as with the 'Asian' in MA-1 but absent in Afontova, which is possibly Oceanic based on some analyses by Raghavan).
Tianyuan is also close to the root of divergence for modern populations, so it makes sense he was somewhat similar to other populations. If we consider a scenario where only ~60% of his genomic diversity survives in modern East Asians, then the rest of his diversity may resemble no modern population and be distributed randomly, or, if Tianyuan's lost diversity was inherited from a common ancestor of him and other populations, then that diversity may have survived in those other populations; hence, he can appear admixed even if he was not. If that diversity was lost from all OoA populations but survives in similar form in Sub-Saharans, then the connection may well lead to an 'African' percentage (which many ancient samples show).
And then there's the problem of low numbers of SNPs and damage which can also seriously confound the results. Pretty much all the ancient DNA analyses I've seen have had membership in at least one inexplicable category, and these often disappear at other k-levels, so some of the stuff that shows isn't always real.
@Tobus
ReplyDeleteUsing the term "Amerindian" in reference to Malta is not misleading just like it's not misleading to talk about "African Americans", although you may want to call blacks "Post-1492 Amerindians."
There's a difference between the data presented by Raghavan and the opinions expressed in that paper. I was referring to the objective evidence, not the subjective interpretation of this evidence by Raghavan.
The only reason you are not seeing evidence for Amerindians at 24000 Ybp in Siberia is because you are denying the Malta evidence for it. You must be also doubting the reality of Denisovans because we only have ancient DNA evidence for their existence in Siberia. You admit that Amerindians are the only living descendants of that ancient population but you refuse to admit that they migrated to Siberia out of America, although it's precisely in Siberia that Malta is located.
In a word, you clearly can't shake off the old myth of Amerindians being the "twelve lost tribes of Siberia" and actually trust the scientific data in front of you.
Finally, I don't have an anti-Asian bias. It's the data that has failed to show East Asians in Siberia at 24,000 Ybp. If Malta had been BLUE and YELLOW, that would have meant that Amerindians who are ORANGE we're not in Siberia at 24,000 YBP because they likely branched off from an admixed west eurasian/East Asian or unadmixed East Asian population in more recent times. But the data is decisively refutes this scenario.
@Hamar Fox
ReplyDeleteActually, all of Eastern View's objections to Raghavan are valid and the fact that you argued for a dual origin of Amerindians before Raghavan simply make you wrong for longer than Raghavan.
There are in fact three Amerindian components in Raghavan: ORANGE, PINK and YELLOW. You can see at K = 3 that YELLOW is Karitiana as much as it is Han. All three components are found outside of America. They are opposed to the BLUE and PURPLE (Papuan) components at the most western and most southern margins of Eurasia. Malta is admixed between west Eurasians, Papuans and Amerindians.
Thank you Hamar Fox, and thank you that you explained the ambiguities in the interpretation of the results!
ReplyDeleteIs it possible to compare Tianyuan and Mal’ta as follows:
Tianyuan Mal’ta
35.39% Veddoid (“South_Asian”) South Asian 37%
31.27% Sinid (“East_Asian”) East Asian 0%
11.32% Indianid (“Amerindian”) Amerind 16%
8.38% Nordic (“North_European”) European 34%
4.84% Tungid (“Siberian”)
3.85% Melanesid (“Australasian”) Oceanian 0.4%
0.00% Eskimid (“Arctic”) Arctic 1%
It is interesting that both Tianyuan and Mal’ta have a similar high percentage of South Asian! There is not a big difference between the Amerind percentages, but Mal’ta seems to be more Amerind than Tianyuan. I would expect that to mean that Tianyuan and the population behind Mal'ta Native American components both contributed genes to Native Americans, and East Asians did not contribute more. I would have expected Tianyuan to be more Tungid. Does that mean that the Tungid component expanded to a bigger extent only after the Ice Age and the original population was small 20-30 kya.
If we go down to just three components, Asian, African, European, I suppose that we cannot any more talk about East Asians, Indians or North Europeans, as this level goes so deep in history that for example whole K may be Asian and GIJ West Asian and E African.
When you say that ”be related on a deeper level with MA-1, regardless of admixture, as with N Europeans, S. Europeans and W. Asians”, I do not quite understand this deeper level relatedness with W. Asians, as there is no West Asian in Mal’ta. Moreover, yDNA P is closer to NO than West Asian J, and mtDNA U is closer to East Asian B than West Asian N1a.
Terry, when you say that “B 'is widely attested in the Americas' but is also widely attested through South China, Southeast Asia and the Pacific islands”, I must add that I checked on Wikipedia B4b frequencies in Asia, and found out that the highest frequencies of B4b, ancestor of Amerind B2, were found in Evenk (New Barag Left Banner) 4/47, Tujia (Yanhe County, Guizhou) 3/29, Mongol (New Barag Left Banner) 3/48 and Han (Beijing) 2/40. Here you have their yDNA C3 frequencies:
West Evenks 28% C3c
Okhotsk Evenks 63% C3c
Outer Mongolia Mongols 34% C3, 19% C3c
Beijing Han 6% C3
Of course, the sister haplogroup of Amerind B2, B4b1, has a more Eastern distribution, being found for example in Japan, Korea, Taiwan and Philippines.
Dziebel,
ReplyDeleteActually, all of Eastern View's objections to Raghavan are valid and the fact that you argued for a dual origin of Amerindians before Raghavan simply make you wrong for longer than Raghavan.
But I've never been wrong about anything in my entire life. Let's both be extremely childish.
There are in fact three Amerindian components in Raghavan: ORANGE, PINK and YELLOW. You can see at K = 3 that YELLOW is Karitiana as much as it is Han. All three components are found outside of America. They are opposed to the BLUE and PURPLE (Papuan) components at the most western and most southern margins of Eurasia. Malta is admixed between west Eurasians, Papuans and Amerindians.
As I said above, ADMIXTURE often gets confused at k=3 regarding Amerindians. I linked to two calculators that allocate Amerindian ancestry differently at k=3 than in Raghavan's runs. That makes ADMIXTURE's say on the matter inconclusive. Therefore, we turn to other measures, as Raghavan did, to draw the most likely conclusion.
Btw, I'd prefer not to be drawn into the ongoing conflict, since I've already concluded it would be fruitless to be so and had deliberately avoided it up to this point.
@German:
ReplyDeleteUsing the term "Amerindian" in reference to Malta is not misleading just like it's not misleading to talk about "African Americans"
But it *is* misleading to say that a 5000yo Ethiopian is "part-African American". Saying Mal'ta is part-Amerindian is putting the cart before the horse, they share some common ancestry, but Mal'ta had that ancestry long before Amerindians were around.
The only reason you are not seeing evidence for Amerindians at 24000 Ybp in Siberia is because you are denying the Malta evidence for it
I see evidence for Amerindian *ancestors* in Siberia at 24kya, but Amerindian haplogroups have an MCRA at ~15kya, and it would be a disingenous to refer to a population before it's own MCRA.
In a word, you clearly can't shake off the old myth of Amerindians being the "twelve lost tribes of Siberia" and actually trust the scientific data in front of you.
I've never heard that myth. The scientific data in front of me tells me there was nobody in America at the time of Mal'ta, and that Amerindians became a discrete population too late to have contributed genes to Mal'ta. What I don't trust is your interpretation that relies on a lot of speculation and very little fact.
It's the data that has failed to show East Asians in Siberia at 24,000 Ybp. If Malta had been BLUE and YELLOW, that would have meant that Amerindians who are ORANGE we're not in Siberia at 24,000 YBP
You are talking like the populations 24 thousand years ago are contemporaneous with modern ones. Assigning modern population components to ancient DNA is a bit like trying to define a primary colour using only tertiary ones (blue is mainly aqua with 30% maroon and a hint of turquiose etc). Don't let the ADMIXTURE analysis confuse you - admixture goes from ancient->modern, not the other way around.
Hamar Fox,
ReplyDeleteI have seen all the professionally done admixture analysis and most of the ones done by highly informed individuals. I don't know what your doc is based out of, so I have no interest in pursuing it.
"They're closest to Mal'ta because they received direct admixture from a Mal'ta-like population. There are two ways a population can share drift with Mal'ta: have direct admixture with his clan (or a similar one), as is true of Amerindians, NE Siberians, and Siberians in general (in that order); or be related on a deeper level with MA-1, regardless of admixture, as with N Europeans, S. Europeans and W. Asians, in that order."
Hamar Fox,
It's well known the Mari, Siberian, and Aleut ethno-history obviously contradicts that, if you ever bother to learn about this populations.
For example, the Aleut having their paternal lines replaced by European males is a historically documented event.
All this is pretty obvious if look at the cluster analysis.
I'm not in the time to go on with the arguments. It's all very parochial.
MY finding is large I found a 17,000 year old y-chromsome from siberia that had Q1a1 and was almost certainly not contamination, because it was published. And people are still arguing over, whether or not malta had american admixture or not. My finding deserves a whole new section. Q1a1 and R* found in paleolithic siberia would be a decent title.
ReplyDeleteEastern View,
ReplyDelete1. But it could not. Never, in this software and with so many comparison data could an individual form its own cluster until you get into the hundreds in K - which is of course meaningless.
2. That interpretation is 100% supported by his y-DNA haplogroup and archeology.
4. Why is that a paradox and not exactly what one would expect?
I don't get your derived logic statement. No one is "pure" - some people are just less admixed since ~35kya, and extant Siberians surely are admixed. Extant Uralic people less so, but still are, even visibly.
Eastern View
ReplyDeleteHamar Fox,
I have seen all the professionally done admixture analysis and most of the ones done by highly informed individuals. I don't know what your doc is based out of, so I have no interest in pursuing it.
That's surprising. I expected you to be highly open to opposing evidence. And that's sarcasm, btw.
One calculator was created by Dienekes, and the other by the owner of the Harappa project. Neither one is incompetent and neither one is dishonest. The ADMIXTURE program is just the ADMIXTURE program. Its algorithms work things out on the basis of the samples it works with. Neither the majority of 'highly professional' people (whom, incidentally, you dismiss when they produce conclusions you personally dislike anyway) didn't develop the ADMIXTURE program any more than did the bloggers who also use it, so your point is garbage.
Also, Raghavan & co. clearly didn't consider ADMIXTURE evidence more compelling than, or even as compelling as, other modes of analysis, which is why they came to the conclusions they did.
Hamar Fox,
It's well known the Mari, Siberian, and Aleut ethno-history obviously contradicts that, if you ever bother to learn about this populations.
Very compelling. I don't think I'll ever fully recover from this crushing blow.
For example, the Aleut having their paternal lines replaced by European males is a historically documented event.
All this is pretty obvious if look at the cluster analysis.
Yes, admixture events specific to Aleuts explain trends that affect all Siberians. Good point. Try to be a little less vague next time, though. You're too crystal clear for my liking.
I'm not in the time to go on with the arguments. It's all very parochial.
Well, it was a very pleasant chat while it lasted.
'Try to be a little more vague' that should be. In the end, I just couldn't out-snark the snarkmeister after all.
ReplyDeleteDaniel Szelkey,
MY finding is large I found a 17,000 year old y-chromsome from siberia that had Q1a1 and was almost certainly not contamination, because it was published. And people are still arguing over, whether or not malta had american admixture or not. My finding deserves a whole new section. Q1a1 and R* found in paleolithic siberia would be a decent title.
Yeah, sorry. I'd planned to address your and Eurologist's posts together. Hardly anything is said about Afontova Gora in either the main paper or the supplementary info, mostly because of his high contamination. There's an interesting PCA in the SI showing him cluster near MA-1, but a little bit closer to the C/S Asian and European clusters than him. The k=4 analysis I've seen (on genetiker's blog) supports this, with him scoring percentages pretty much exclusively in the 'European' and 'Amerindian' categories (at roughly 70% European and 30% Amerindian), while MA-1 also had around 7% in the Asian column. That maybe suggests that MA-1's population isn't ancestral to AG's, but is obviously closely related. It also suggests that there wasn't much of a migration back into Europe by these Siberians, at least not paternally, since their Y-DNA is rare to absent further west.
The really interesting question is whether the Q mutation occurred between MA-1's time and AG's, or these two similar populations sat side by side in Siberia for the longest time before Q migrated East, absorbing MA-1-like populations as they went.
@hamer fox
ReplyDeletehttp://www.yfull.com/full-genomes/
AF2 is Afantova=2
the paper says nothing about afantova y-chromosome
but y-full is a offical reliable source: and says Afantova belongs to Q1a1-F1215.It is not publisheed on nature it is published on y-full.
Q1a1-F1215 is ancestral to the type of Q-most common in Turkmens it has the main clade of Q, in east and central asia. It is rare in america, wher Q1ab dominates, I think this suggests Q is at least 30,000 kya and is probably a little older then R,
Mal'ta is also on this page:however, 7KYA is a long time.
Y-FULL also notes Afangora's mtdna which is a unresolved or undisclsoed clade of macro haplogroup R.You can also find malta on this page by searching for MA-1. Mal'ta and Afonogora are two are the four cases reported in popualtion none.
There was again an error in my comment: Mal’ta Oceanian component is probably 4% and Arctic component 10%?
ReplyDeleteIt is useless to argue over opinions that cannot be proven at the moment, but according to Wikipedia http://en.wikipedia.org/wiki/Models_of_migration_to_the_New_World the earliest date for the settlement of Beringia by the ancestors of Native Americans is 36 kya. The oldest clades of haplogroup A are: A2, America, 20 kya, A4b, Okhotsk Sea area, Inner Mongolia, Ob river area 15 kya, A5b, Altai, Japan, China, 17 kya and A8, Yenisei area, Kamchatka, 18 kya. The age estimation of B4b, ancestor of Native American B2, found for example in Tujia, Evenks and Mongols, is 28 kya. The oldest clades of haplogroup C are: C7, India, Siberia, 26 kya, C4a, Siberia, India, Tibet, China (?), 25 kya and C1, America, 21 kya. The age estimation of Native American D1 is 17 kya, and recently one Peruvian mummy fell into haplogroup M (an ancestor of D).
On the basis of this, I think that it is not at all impossible that the ancestors of Native Americans were already in Beringia or in the area between Beringia and Lake Baikal, or Beringia and North China 24 kya.
@Daniel Szelkey,
ReplyDeleteIf I am not mistaken, they also found a marker for R1a1a1 at the M417 level, but as that would make it inconsistent with current calculated ages for R1a1a1, so it is thought to be from contamination as the sample quality is very bad. But there are no positive SNPs below M417 level.
@Tobus
ReplyDeleteYou are using a wrong analogy. Imagine we would find ancient DNA in Eurasia with an African component in it. We would immediately interpret it as a signal of an out of Africa migration. The Malta situation is no different. The problem is that we don't see a trace of Africans in this ancient Eurasian find. We find Amerindians where we would expect Africans under the out of Africa model.
Regarding the "twelve lost tribes of Siberia", I thought you would appreciate the irony of having an old European idea of Amerindians being the "twelve lost tribes of Israel" reappear with Siberia as the mythical source.
I've neglected to answer your repeated reference to y DNA and mtdna trees and associated dates because I don't want to open a can of worms. Suffice it is to say that they date to pre ancient DNA and pre ADMIXTURE kind of analyses and need to be revised accordingly. Molecular clock is notoriously fishy and shouldn't be used to trump archaeological dates.
I didn't understand your color analogy. ADMIXTURE is the analysis widely applied to modern populations and only incipiently to ancient ones.
"The scientific data in front of me tells me there was nobody in America at the time of Mal'ta, and that Amerindians became a discrete population too late to have contributed genes to Mal'ta. What I don't trust is your interpretation that relies on a lot of speculation and very little fact".
ReplyDeleteAgreed.
"Does that mean that the Tungid component expanded to a bigger extent only after the Ice Age and the original population was small 20-30 kya."
I would think that is the case. But not only was the 'original population' small, from this paper we see it also had very little of any 'East Asian' element at first. Y-DNAs N and C3 were almost certainly involved in later expansions north, although members of C3 were obviously early enough to enter America before N expanded.
"I checked on Wikipedia B4b frequencies in Asia, and found out that the highest frequencies of B4b, ancestor of Amerind B2, were found in Evenk (New Barag Left Banner) 4/47, Tujia (Yanhe County, Guizhou) 3/29, Mongol (New Barag Left Banner) 3/48 and Han (Beijing) 2/40".
That is completely expected, surely. Haplogroup 'B4b, ancestor of Amerind B2,' is in fact the main B clade found outside South China/SE Asia. B4b belongs within a clade that includes B4d, B4e and B4j. Of these others just the last can be considered at all 'north' Asian. B4b'd'e'j belongs to a clade that includes B4f, B4c, B4g, B4i and the large B4a group. Basically none of these are present in the north. And B4 is a 'sister' to B5, basically Laos, Vietnam and Sumatra although B5a1 reached as far north as Japan. Further back still B4'5 is 'sister' to both R24 and R11'B6. The former is confined to the Philippines, the latter in Yunnan, Burma and Laos. That makes the whole haplogroup almost certainly southern in origin.
"Of course, the sister haplogroup of Amerind B2, B4b1, has a more Eastern distribution, being found for example in Japan, Korea, Taiwan and Philippines".
And even as far out into the Pacific as the Admiralty Islands evidently. It would almost certainly have arrived there from Taiwan and the Philippines with Austronesian B4a1. Many suspect there was some influence from Japan at the beginning of the Austronesian expansion and so perhaps B4b1 was part of that. Thanks for the yDNA C3 frequencies.
It is interesting that both Tianyuan and Mal’ta have a similar high percentage of South Asian!
ReplyDeleteKristiina,
That's roughly where almost all extant ooA people originate from, so the deeper you go in time, the more you will see this, since obviously extant S Asia has least drifted from ancient S Asia.
When you say that ”be related on a deeper level with MA-1, regardless of admixture, as with N Europeans, S. Europeans and W. Asians”, I do not quite understand this deeper level relatedness with W. Asians, as there is no West Asian in Mal’ta.
I can't speak for Hamar Fox, but I would support the same. N Europe (including N Iberia) was during the Gravettian affected by a wave (R1a, R1b?) probably very similar to the Q and R* waves in Siberia. S Europe (excluding Iberia) was less and differently affected during the Gravettian, but still shares the Aurignacian with N Europe. The southern portion of W Asia (roughly, Anatolia and south) appears to have been populated by more southerly NW subcontinent people (y-DNA J and G, and later T, L, etc.) but shares at least some of the Aurignacian (IJ?) and the general NW subcontinent origin with Europe and Siberia.
"West Asia" as a component had ~50,000 years to differentiate, with strong remaining contact to "Gedrosia" and S Asia. N Europe only had 20 to 30 ky to differentiate with Siberia, with likely strong remaining contact (at least diffusion) during the entire time (remember that S Siberia was pretty much continuously and contiguously settled throughout even LGM!).
Tianyuan, 40 kya, has 31.27% of Sinid (East Asian) and 4.84% of Tungid (Siberian). According to the Chinese Super-Grandfathers paper, the divergence between C3 and C(xC3) happened 40 kya and the divergence between P and NO 33 kya. If O is expected to have contributed to Tiayuan, the age estimations must be too low. As, according to the same Chinese paper, the divergence between Q and R happened 24 kya and we know now that Mal’ta was hg R already 24 kya, these estimations should be scaled upwards. The big question is of course how much!
ReplyDeleteWhen we take into account what Maju says on his blog that “the fact that the admixture signal stems from quite upstream of MA-1 indicates that this boy ... was not direct ancestors of Native Americans in any significant way but rather a different branch from the same trunk”, the interesting thing is that Mal’ta is however more Amerind (16%) than Tianyuan (11.32%). Then, if we consider the possibility that yDNA R came from north of India and mtDNA U from Europe, Mal’ta boy must have mixed with a population very high in Amerind component. One could even calculate that these paleo-Native American relatives of Mal’ta were 88% Amerind.
Note also that the other Native American component is pink “Arctic” one, and Mal’ta is 10% Arctic, while this component is absent in Tianyuan!
On the basis of this, Native Americans and Arctic people seem to be a very different set of people from Han Chinese and other Southern Mongoloids.
"I think that it is not at all impossible that the ancestors of Native Americans were already in Beringia or in the area between Beringia and Lake Baikal, or Beringia and North China 24 kya."
ReplyDeleteExtremely likely. I think the evidence as we have it suggests Y-DNA Q and mt-DNA X were a sort of 'advance guard' for the movement east from the steppe. Y-DNA R and mt-DNA U followed along behind and replaced Q and X through much of their earlier range. However the Q/X and R/U populations had each originally been part of the same west Eurasian population, as represented by the Mal'ta boy.
"The Malta situation is no different. The problem is that we don't see a trace of Africans in this ancient Eurasian find. We find Amerindians where we would expect Africans under the out of Africa model".
By the time of the Mal'ta boy the OoA event is long past. And we don't 'find Amerindians' in 'this ancient Eurasian find'. We find an element common to west Eurasians and Amerindians. As I keep trying to point out, the Mal'ta boy and Amerindians share just 14 to 38% of their aDNA. You cannot seriously claim the Mal'ta boy is 'Amerindian'.
"I've neglected to answer your repeated reference to y DNA and mtdna trees and associated dates because I don't want to open a can of worms. Suffice it is to say that they date to pre ancient DNA and pre ADMIXTURE kind of analyses and need to be revised accordingly".
You seem still to believe that admixture alters haplogroups. What do they admix with? The advantage of haploid genes over diploid is that they alter only as a contained series of mutations. Diploid genes keep mixing each generation as well as mutating over time. They are far more complicated to unravel.
"Molecular clock is notoriously fishy and shouldn't be used to trump archaeological dates".
I agree the 'molecular clock' is not particularly accurate but molecular clock doesn't come in to consideration of haplogroup phylogeny.
"As, according to the same Chinese paper, the divergence between Q and R happened 24 kya and we know now that Mal’ta was hg R already 24 kya, these estimations should be scaled upwards. The big question is of course how much!"
ReplyDeleteAn example of the unreliability of the molecular clock, as in German's comment above.
"this boy ... was not direct ancestors of Native Americans in any significant way but rather a different branch from the same trunk"
Yes. As I said above he is part of a slightly later movement across the steppe from the 'ame trunk'.
"the interesting thing is that Mal’ta is however more Amerind (16%) than Tianyuan (11.32%)".
Because they originate in different places. Mal'ta from the west Eurasia steppe and Tianyuan from southern China. Basically the Mal'ta boy carries the west Eurasian element of the Amerindians and Tianyuan the East Asian element.
"Then, if we consider the possibility that yDNA R came from north of India and mtDNA U from Europe, Mal’ta boy must have mixed with a population very high in Amerind component".
No. The Mal'ta population, or something very similar, mixed with an East Asian population that became Amerindian.
"On the basis of this, Native Americans and Arctic people seem to be a very different set of people from Han Chinese and other Southern Mongoloids".
The two groups have mixed with different populations. In the north amixed with a 'Mal'ta-type' population (followed by further movement north of an East Asian population), and admixed in the south with a Papuan/Australian-type population. This has led to the formation of 'northern' and 'southern' Mongoloids.
@terryt
ReplyDeleteADMIXTURE shows at k = 2 two components: Amerindian and African. (Hamar Fox should toss away any other muddled runs he got his hands on.) it means you can build a tree with basal branches in America or in Africa. Current mtdna phylogenies have their haplotypes built without any awareness if the ancestral states as ascertained in Neanderal and Denisovan sequences. Your favorite hg B2, for example, has a number of ancestral retentions that PhyloTree miscodes as innovations. Same for L3 and N. So I wouldn't use the phylogeny as your hotel bible. It needs to be revisited with the use of ancient DNA.
"You can't seriously claim that the Malta boy is Amerindian."
We should ask the boy himself. Hundreds of people in the US have that amount of Amerindian "blood" (the very same component!) and they call themselves Amerindian. Also Malta is the closest to Amerindians among all living populations, according to one of the charts in Raghavan. A clear back flow from the Americas if you dump your biases. We can call the boy "Amerindian Siberian."
I couldn't get this posted for some reason before. Hopefully it works this time.
ReplyDeleteFor what it's worth...
eurogenes v2 k15 average for NorthAmerindian
North Sea .27
Atlantic .09
Baltic .3
Eastern Euro 2.27
West Med .01
West Asian .07
East Med .01
Red Sea .07
South Asian 1.7
Southeast Asian 5.08
Siberian 20.17
Amerindian 69.81
Oceanian .11
Northeast African .03
Sub Saharan .02
The main carriers of the Arctic component are Koryaks, Chukchis and Greenlanders. Their yDNA frequencies are the following (omitting western haplogroups of Greenlanders, including R):
ReplyDeleteKoryaks: Q 18.5%, N1c 22.2%, C3 25.9%, C3c 33.3%
Chukchis: Q-M3 12.6%, R1a 4.2%, P(xR) 20.8%, N1c 58.3, C3c 4.2%
Greenlanders: C 1/69, Q(xM3) 9/69, Q-M3 4/69
Only haplogroups Q and C are found in all groups. According to the admixture analysis, Greenlanders lack Siberian, light yellow component, while Koryaks and Chukchis carry it at a frequency of c. 30%, and all three groups lack darker yellow, East Asian component. N1c’s (note, not East Asian N-LLY22g's) connection to the introduction of Siberian element in Koryaks and Chukchis is very plausible. Taking into account the age of N1c, this expansion probably occurred less than 10 kya. If C3c is part of the original Arctic component together with Q(xM3), it is interesting that the Arctic component is absent in Tianyuan (40 kya) and found in Mal’ta (24 kya) at a frequency of 10%. Is it then possible that the ancestor of C3c arrived from Altai to North China c. 30 kya spreading to the North East and mixed with a group related to Mal’ta boy near Baikal area? This population that passed through Northern China might have then taken B2 to America and introduced mtDNA A to China.
MDLP K27 admixture of the Mal'ta boy
ReplyDelete0.001 Nilotic-Omotic
4.6573 Ancestral-South-Indian
11.4993 North-European-Baltic
38.4626 Uralic
0.001 Australo-Melanesian
0.001 East-Siberean
0.001 Ancestral-Yayoi
0.001 Caucasian-Near-Eastern
0.001 Tibeto-Burman
0.001 Austronesian
0.001 Central-African-Pygmean
0.001 Central-African-Hunter-Gatherers
0.001 Nilo-Saharian
0.001 North-African
25.3658 Gedrosia-Caucasian
0.001 Cushitic
0.001 Congo-Pygmean
0.001 Bushmen
0.001 South-Meso-Amerindian
0.001 South-West-European
17.3835 North-Amerindian
0.001 Arabic
0.001 North-Circumpolar
0.001 Kalash
1.1879 Papuan-Australian
0.001 Baltic-Finnic
1.4236 Bantu
@German:
ReplyDeleteYou are using a wrong analogy
It was *your* analogy. This is the 2nd time you've presented something to me and then told me I shouldn't be using it. Please stop doing this - if you're wrong just admit it, don't try to make out it was my argument.
I didn't understand your color analogy
It's pretty simple. Tertiary colours like turquoise and maroon come before primary colours like blue - you can can make turquiose and maroon using blue, but it's folly to try to make blue by mixing blue-derived colours like turquoise and maroon. The analogy is with modern and ancient populations in ADMIXTURE. All the ADMIXTURE components are from modern populations and can be described as being mixtures of earlier populations. MA-1 however is an ancient population, so it's folly to claim it's a mixture of any modern populations - MA-1 came first, modern populations second. What the analysis tells us is which ancestors MA-1 shared with modern populations, not which modern populations mixed to create MA-1. Modern populations (ie Amerindians) are mixtures of ancient populations... but ancient populations (ie MA-1) can only be mixtures of even more ancient populations. Your notion that MA-1 is "part-Amerindian" is incorrect.
ADMIXTURE shows at k = 2 two components: Amerindian and African
Only if "Amerindian" is redefined to mean "non-African". You are confusing the colours on admixture at the lower K levels with the final colours at higher levels. Quite hypocritically too since you are more than willing to say the opposite when it suits you - eg " Yellow is just as much an Amerindian component as it is East Asian" (so vice versa, orange is as much an East Asian component as it is Amerindian).
We can call the boy "Amerindian Siberian."
What's wrong with "Ancient Siberian"? It's only *your* biases that makes you want to include "Amerindian".
This comment has been removed by the author.
ReplyDelete@Tobus
ReplyDelete"... a 5000yo Ethiopian is "part-African American" was your analogy. What is wrong with you? You're trying to engage in high scholarly debates but I need to correct you on simple things.
Once again: imagine we find a 25,000 YO DNA from India that carries an "African" component. Would you be arguing that because it's 25,000 years old and it was found outside of Africa, it describes a population ancestral to modern Africans? Of course, not. You'll be claiming that it's a trace of an out-of-Africa migration.
"MA-1 however is an ancient population, so it's folly to claim it's a mixture of any modern populations - MA-1 came first, modern populations second. "
I see that logic is not your best friend. Modern populations haven't sprung from the breath of God. They represent generations of unbroken descent. In the case of the Amerindian component it goes back to at least 25,000 years and since it's ascertained all over America at high frequencies and not as much in Siberia, it's an "Amerindian component" that entered Siberia 25,000 years ago and spread westward far and wide.
With your bizarre logic, West Eurasians, Amerindians, Papuans and South Asians all originated in Siberia from a Mal'ta-like population.
As I told you already, if you want to build an ancient DNA case for the origin of Amerindians from Asia, you need to have a Paleoindian DNA with an East Asian component in it.
"Only if "Amerindian" is redefined to mean "non-African"."
Another bias. Why "non-African"? Amerindian vs. African is fine. It'll be even more accurate to say that worldwide variation is bookended by Sub-Saharan Africans and Sub-Rio Grande Amerindians, with all other populations falling in between.
"You are confusing the colours on admixture at the lower K levels with the final colours at higher levels."
Raghavan's run is incomplete as it doesn't include K=2. That's why I brought up Rosenberg et al. 2002, so you can see a complete picture. Raghavan's K=3 is confusing because it paints Amerindians YELLOW but they just have as much yellow as East Asians. Mal'ta DNA results wouldn't make any sense if Amerindian ORANGE was derived from East Asian YELLOW as this would make Mal'ta as close to East Asians as they are to Amerindians. The latter is demonstrably not true: Mal'ta and other west Eurasian populations are closer to Amerindians that to east Asians (see Figure SI 25, SI 26, SI 22, SI 9, etc.). I finally got back and revisited the data: your claim that Amerindians are closer to East Asians than to Mal'ta is too simplistic and is contradicted by a host of charts in Raghavan et al.
The only possible interpretation is that Amerindians are not "admixed" but "diverse and sub-divided." They furnish a common denominator between East Asians and West Eurasians. Africans, on the other hand, are likely heavily admixed but equilibrated so that ADMIXTURE interprets it as a single component.
This has been my consistent position for over 10 years, since I saw heard Marcus Feldman present Rosenberg's novel approach, and I honestly don't know how you can call it "hypocritical."
"What's wrong with "Ancient Siberian"? It's only *your* biases that makes you want to include "Amerindian"."
How in the world can it be a bias if IT IS an "Amerindian component"? "Ancient Siberian" as a name for the Mal'ta population is like "new American" as the name for "African Americans" - empty at best, weird at worst.
@Eastern View
ReplyDelete"MA-1 is allowed to form its own cluster, and then have other samples be derived from it, which is the most natural way to determine anything."
Bingo!
This comment has been removed by the author.
ReplyDelete@Eastern:
ReplyDelete"MA-1 is allowed to form its own cluster, and then have other samples be derived from it ... Mal'ta should form it's own cluster if it is "pure"
I'm not sure how this relates to what I'm saying - you can't claim admixture goes backwards in time. Regardless of how he clusters, no modern populations contributed any DNA to Mal'ta, all his DNA came from people who existed before him, not after. Shared components means shared ancestry or admixture from Mal'ta, never admixture to Mal'ta from a modern group.
MA-1 is allowed to form its own cluster, and then have other samples be derived from it, which is the most natural way to determine anything. It does not form its own cluster. In comparison, a single sample of, say, the Paniya, shows that it mostly belongs a green "Äncestral South Asian" cluster. There is really no difference between MA-1 and Paniya. Time or number of samples matters not. What matters if a pattern is integral compared to a host of other samples. Mal'ta should form it's own cluster if it is "pure".
ReplyDeleteYour Paniya example has no basis. The Paniya are represented by four samples in the ADMIXTURE analysis, not one, and more importantly, there are other Indian populations besides the Paniya in the ADMIXTURE analysis, each one of whom is represented by multiple samples, and their ADMIXTURE results are similar to those of the Paniya, thus in the ADMIXTURE analysis the Paniya are by no means the sole representative of Paniya-like populations. In contrast, in the ADMIXTURE analysis the MA-1 sample is the sole representative of MA-1-like populations; so no wonder that no MA-1-specific component emerges in the ADMIXTURE analysis. If a few more MA-1-like samples were available, a MA-1-like-specific component would surely emerge.
In addition, the green component you mention is not an ASI component but a composite of Caucasoid and ASI or ASI-like ancestries, and it is made up mostly of Caucasoid ancestry in South Asian populations and almost exclusively of Caucasoid ancestry in West Eurasian populations.
@German:
ReplyDeleteimagine we find a 25,000 YO DNA from India that carries an "African" component. Would you be arguing that because it's 25,000 years old and it was found outside of Africa, it describes a population ancestral to modern Africans
No, because there were lots of people living in Africa long before 25kya, and Africans have a MCRA long before 25kya. You are presenting a false analogy to the Mal'ta situation because there were no Amerindians around that Mal'ta could be the ancestor of, but plenty of Africans to be the ancestor of your hypothetical Indian.
A much better analogy would be finding "Eurasian" DNA in a Northeastern African population from a few thousand years before the Out Of Africa migration. By your logic this proves there were Eurasians in Africa and Africans are part-Eurasian. You ignore that "Eurasian" didn't exist as a population before the OOA event, and that the "first Eurasians" had, geographically speaking, 100% African DNA. Their ancestors and relatives who stayed in Africa shared much of that DNA - we only call it "Eurasian" now because all present-day Eurasians have this group as their MCRA and it has disappeared almost completely in Africa. The same applies to Amerindians, the "first Americans" went to America with, geographically speaking, 100% Eurasian DNA and their ancestors and relatives who stayed in Eurasia (Mal'ta being a distant example) shared much of this DNA. We only call it "Amerindian" now because all present-day Amerindians have this group as their MCRA and it has disappeared almost completely from Eurasia.
With your bizarre logic, West Eurasians, Amerindians, Papuans and South Asians all originated in Siberia from a Mal'ta-like population.
No, you have confused yourself again. Mal'ta pre-dating modern populations doesn't mean he is the direct or only ancestor of all modern populations - it just means that no modern populations contributed anything to his DNA, his DNA all came from his ancestors, some of which are also ancestors of some modern populations. It's funny you find such a concept bizarre though, woldn't this be exactly the scenario an Out Of America theory suggests - Americans (and hence Siberians) being the ancestors of every modern population on the planet?
Another bias. Why "non-African"? Amerindian vs. African is fine
"Non-African" because the K=2 colours are split between African and non-African populations. "African vs Amerindian" ignores the non-Amerindian populations included in the same colour.
if you want to build an ancient DNA case for the origin of Amerindians from Asia, you need to have a Paleoindian DNA with an East Asian component in it.
... as you already said "Yellow is just as much an Amerindian component as it is East Asian", so we already have it.
Raghavan's K=3 is confusing because it paints Amerindians YELLOW but they just have as much yellow as East Asians
... and you just said it again!
Mal'ta DNA results wouldn't make any sense if Amerindian ORANGE was derived from East Asian YELLOW as this would make Mal'ta as close to East Asians as they are to Amerindians
No it wouldn't... and especially not if the orange contains some non-yellow admixture.
your claim that Amerindians are closer to East Asians than to Mal'ta is too simplistic and is contradicted by a host of charts in Raghavan et al.
Which ones? Not the ones you refer to above I assume, since SIs 26 and 9 show Amerindians are closer to East Asians than they are to Mal'ta and SIs 25 and 22 don't measure affinity between the two.
(cont.)
ReplyDeleteI honestly don't know how you can call it "hypocritical."
I find it hypocritical that you combine East Asians and Amerindians when it suits you, but not when it doesn't... do you accept that the "Amerindian" on Rosenberg's chart could just as easily be called "East Asian"?
How in the world can it be a bias if IT IS an "Amerindian component"
Mal'ta is primarily South Asian and European components, but no one is you are not calling him "Indian Siberian" or "French Siberian". You want to imply that modern-day Amerindians existed and were in Siberia at 24kya so you emphasise the "Amerindian" component out of proportion. That's your bias showing.
"Ancient Siberian" as a name for the Mal'ta population is like "new American" as the name for "African Americans" - empty at best, weird at worst.
OK, let's just call him "Mal'ta" then.
"I see that logic is not your best friend. Modern populations haven't sprung from the breath of God. They represent generations of unbroken descent. In the case of the Amerindian component it goes back to at least 25,000 years and since it's ascertained all over America at high frequencies and not as much in Siberia, it's an 'Amerindian component' that entered Siberia 25,000 years ago and spread westward far and wide".
ReplyDeleteFirst off, most populations do not 'represent generations of unbroken descent'. They represent generations of isolation often followed by mixture with other populations. That's why it can be said there are no pure populations. I agree that the 'Amerindian component' entered Siberia 25,000 years ago and spread westward far and wide but the current paper tells us that the 'Mal'ta' population provided just a proportion of that 'Amerindian component'. Amerindians usually show a connection to East Asian populations and so we can safely assume that this population, not present in the Mal'ta boy, makes up the non-Mal'ta element of the Amerindian component. I would have though that was very simple to comprehend.
"It'll be even more accurate to say that worldwide variation is bookended by Sub-Saharan Africans and Sub-Rio Grande Amerindians, with all other populations falling in between".
You're ignoring Australian Aborigines here. I agree very lttle work has been done with them but what work that has been shows they form a pole of variation with the rest of the world.
And I notice you still haven't explained how haplogroups can change simply through mixing with other haplogroups.
@Tobus
ReplyDelete"No, because there were lots of people living in Africa long before 25kya..."
This is an African bias. It doesn't matter what other data says about Africans' antiquity. You need to take every evidence on its merits. Otherwise, circular reasoning ensues.
"We only call it "Amerindian" now because all present-day Amerindians have this group as their MCRA and it has disappeared almost completely from Eurasia."
Another piece of flawed logic. Why would one need to postulate the extinction of a proto-Amerindian population in Eurasia if we have this population well and alive in America? "Living population" doesn't mean "recent population."
"A much better analogy would be finding "Eurasian" DNA in a Northeastern African population..."
I'm so glad you describe your flawed thinking in such a great detail. Of course, if you assume that out-of-Africa happened, then anything that you find in Africa becomes either eternally African or proto-Eurasian. The Hofmeyr skull from 35,000 YBP in South Africa is a good real world example. It clusters with UP Eurasians and not modern Africans or AMH. By a normal logic, it's a Eurasian skull representing a migration into Africa. If we apply to it an African bias, however, then of course Eurasians can be claimed to have originated in South Africa. But that won't be science anymore. Plain ideology.
"Mal'ta pre-dating modern populations ...means that no modern populations contributed anything to his DNA..."
This is profound. So, your contribution to the debate amounts to saying that ancient samples are not derived from modern populations?
"Shared components means shared ancestry or admixture from Mal'ta, never admixture to Mal'ta from a modern group."
Oh, this must be why Raghavan et al. spent one whole page of their main article building an argument that Mal'ta didn't receive gene flow from Amerindians. Apparently, they didn't know that you waived the need to prove it.
"woldn't this be exactly the scenario an Out Of America theory suggests?"
yes, in an exact reversal of the basic out-of-Africa premise. America and Siberia are in fact in a better position because we can have ancient DNA from these regions to test it. Out of-Africa conveniently placed the "origin of man" in the latitudes incapable of generating any ancient DNA material to test it.
"do you accept that the "Amerindian" on Rosenberg's chart could just as easily be called "East Asian"?"
No. Just look at the Rosenberg chart. At K=2 Amerindians and Africans are the only populations that are pure in their respective components. East Asians, Papuans and West Eurasians are a mix of both. Papuans are most Amerindian, East Asians are more Amerindian and West Eurasians are more African. At K=3 east Asians are a mix of West Eurasians and Amerindians. At K=4 through K=6 east Asians are an independent entity, and these independent east Asians are not found in the Americas. Amerindians retain their own color through all the levels.
"no one is you are not calling him "Indian Siberian" or "French Siberian"."
People easily refer to Mal'ta as genetically "West Eurasian" because its Y-DNA and mtDNA are such. What's the problem? I don't call Mal'ta "Siberian Cheyennes" but Amerindian Siberians are a legitimate label, but, admittedly, not the only possible one.
@Tobus (contd.)
ReplyDelete"Which ones?.."
Precisely the ones I listed. Mal'ta and other West Eurasians are closer to Amerindians than they are to East Asians. E.g., "Tendency of Middle Eastern, European, and Central and South Asian populations to all be closer to Native Americans than to Tianyuan" (S25). S22 shows shared drift between Malta and global populations: Amerindian values are the highest and they are "red." From S9 it's clear that Malta is closer to Amerindians than it is to East Asians. (If East Asians predated and generated Amerindians, they would've been closer to Mal'ta than Amerindians.) S26 confirms that "all
western Eurasian populations are closer to Karitiana than to Han."
@Kristiina
"German, you checked hg P and did not find any excess in Denisovan/Neandertal matches. How about putative M29’Q or Australian M15? "
I finally checked, but only the stem substitutions along M29'Q, M29, Q and M15. I didn't go into any subclades and I didn't go upstream. M29 and M15 do look interesting: M29 has some 35% "ancestral retentions", M15 just under 30%. On M29 two sites 151, 152 show the same base as Sima, Denisova 1 and Denisova 2, 152 is a hot spot in modern humans, though; 1048 is the same as Denisova 1, 2 as well as L0. I didn't go that far aligning Sima against other sequences, so I don't know if Sima is C ot T at 1048.
@ terryT
ReplyDeleteFirst off, most populations do not 'represent generations of unbroken descent'. They represent generations of isolation often followed by mixture with other populations."
What kind of idea is that, Terry: populations represent generations of isolation but not descent? Isolation reduces the number of lines of descent but it doesn't destroy descent. And yes of course populations mix with each other. Africans are the most admixed (highest intragroup diversity values globally), while Amerindians are the most isolated (highest intergroup diversity values globally).
"I agree that the 'Amerindian component' entered Siberia 25,000 years ago and spread westward far and wide."
Good, finally.
"the 'Mal'ta' population provided just a proportion of that 'Amerindian component'."
What are you talking about? Mal'ta has other components in addition to the Amerindian one but it didn't provide Amerindians with a special Malta component.
"Amerindians usually show a connection to East Asian populations and so we can safely assume that this population, not present in the Mal'ta boy, makes up the non-Mal'ta element of the Amerindian component. I would have though that was very simple to comprehend."
Where is that "non-Mal'ta component" that Amerindians got from East Asians in the ADMIXTURE graphs in Raghavan? In fact, the best way to think about the data at hand is that Mal'ta is an ancient East Asian population formed mostly by admixture from West Eurasia and the southern areas of the New World (see also the Rosenberg run that I referenced in my exchange with Tobus). A more recent East Asian population, which we associate with the Mongoloid phenotype (EDAR, shovel-shaped incisors, etc.) is likely product of admixture between the already admixed ancient east Asian population and a new wave of northern Amerindians (relatives of Na-Dene and Eskimo-Aleuts) that brought the specific Mongoloid phenotype to East Asia after the retreat of the glacier around 12,000 years ago.
"And I notice you still haven't explained how haplogroups can change simply through mixing with other haplogroups."
I thought I wrote something above in a couple of places. The main takeaway for you is that current mtDNA haplotypes were built in pre-ancient DNA times. Consequently, they miscode a number of ancestral retentions ascertained in Denisova (with or without Neandertals) as innovations. Plus they are full of reverse mutations and homoplasies. In a word, they need to be revisited at a fundamental level. But even if we don't go this route in the nearest future, admixture can shift the root of a tree pretty dramatically because it brings a bunch of ancient sequences into a more recent population thus making it look older than it actually is.
I have some questions here. Excuse me if it is missing the point, or way off base. I am by no means a professional, and just beginning my genetics and anthropology studies.
ReplyDeleteObviously we cannot say that the Mal'ta individual is from modern populations (for instance, 33% Tajik, 66% Udmurt), but can't we possibly see that the various admixture components (Gedrosia, Uralic, Amerind, etc.) are much older. Why would an individual show these populations, if they were not descended from these groups? If these separations had not occurred, why would they appear in say a k27 run? I don't think it's unreasonable to say that these things could be very old, near the East and West Eurasian split. If I am wrong in my assumption, can someone please provide references to their answers (preferably peer-reviewed if possible). Perhaps with more specimens from the area and time-frame we will know more. The Gedrosian percentage has me wondering; could we be seeing someone who is an offspring of a majority Gedrosian parent and a majority Uralic parent? Perhaps it will help us to understand not just the East and West Eurasian migrations, but also component migrations/admixture events.
Just running some thoughts by you all. Please, again provide reasoning for your claims and references, so that I may study and learn more. If it's not too much to ask, let's not have any ad hominem comments back and forth. Thanks, in advance for you input!
@German;
ReplyDeleteWhy would one need to postulate the extinction of a proto-Amerindian population in Eurasia if we have this population well and alive in America
Because that population wasn't well and alive in America at the time of Mal'ta - I thought I already said that.
So, your contribution to the debate amounts to saying that ancient samples are not derived from modern populations?
It's only half my contribution (the other half being that Amerindians are closer to East Asians than to Mal'ta). Your statement that Mal'ta is "part-Amerindian" means that there had to be an Amerindian population pre-dating Mal'ta for him to be descended from, but there wasn't. If taken as a genetic term then "Amerindian" means people descended from the MCRA of modern Amerindians, which, as you know, is dated to ~15kya, long after Mal'ta. If taken as a geographical term "Amerindian" means people physically present in America and (despite your personal speculations) there is no evidence of anybody living in America 24,000 years ago. Unless you want to redefine what "Amerindian" means, Mal'ta can't be "part-Amerindian", it would require a time machine.
Precisely the ones I listed. Mal'ta and other West Eurasians are closer to Amerindians than they are to East Asians
Yes, I agree, but you were supposed to be providing evidence to support the statement "your claim that Amerindians are closer to East Asians than to Mal'ta is too simplistic and is contradicted by a host of charts in Raghavan et al."... so I was expecting charts that show Amerindians are closer to Mal'ta than Amerindians are to East Asians, not charts that show Mal'ta is closer to Amerindians than Mal'ta is to East Asians.
Just to recap, your original statement was "modern Eurasian populations are less Amerindian than Mal'ta" and you appeared to retract that when you said "I originally meant that Malta has more of the Amerindian component than west Eurasians". I have no problem with the 2nd statement, but the first ignores that East Asian/Amerindian affinity is greater than Mal'ta/Amerindian.
@Tobus
ReplyDelete"Because that population wasn't well and alive in America at the time of Mal'ta..."
So you postulate the extinction of a proto-Amerindian population in Siberia and the lack of antiquity of the Amerindian population in America? Well, this is clearly biased and self-serving. Mal'ta proves that Amerindians were around at 25,000 YBP and this is consistent with archaeology and linguistics. See the latest find at 30,000 YBP in Uruguay see (http://rspb.royalsocietypublishing.org/content/281/1774/20132211.full.pdf+html).
"Your statement that Mal'ta is "part-Amerindian" means that there had to be an Amerindian population pre-dating Mal'ta for him to be descended from, but there wasn't."
Mal'ta DNA does suggest that Amerindians predate Mal'ta. It doesn't say, of course, by how much but the other thing it suggests is that East Asians postdate it.
"Yes, I agree, but you were supposed to be providing evidence to support the statement "your claim that Amerindians are closer to East Asians than to Mal'ta is too simplistic and is contradicted by a host of charts in Raghavan et al."... so I was expecting charts that show Amerindians are closer to Mal'ta than Amerindians are to East Asians, not charts that show Mal'ta is closer to Amerindians than Mal'ta is to East Asians."
No, I was supposed to provide evidence that your claim that A. Amerindians are closer to East Asians than to Mal'ta, coupled with the claim that B. There were no Amerindians during Mal'ta times are flawed.
The first one is simplistic and incomplete as Mal'ta and West Eurasians are closer to Amerindians than they are to East Asians, which dramatically changes the meaning of your (factually correct) claim A. West Eurasians are at least 24,000 years old and are not derived from East Asians. This means that if they are closer to Amerindians than to East Asians, Amerindians are, too, at least 24,000 years old.
If East Asians are closer to Amerindians than they are to West Eurasians and West Eurasians are closer to Amerindians than they are to East Asians, it means Amerindians likely predated and spawned at least of these populations. Since East Asians are closer to Amerindians than West Eurasians, East Asians are more likely to be derived from Amerindians than West Eurasians. This is consistent with the presence of the West Eurasian genetic component and the absence of the East Asian genetic component in an East Asian geographic location at 24,000 years.
So your whole argument is thus shown to be flawed. Your claim A was incomplete and claim B factually wrong. You tied them together to produce an utterly distorted picture.
"I thought I wrote something above in a couple of places [how haplogroups can change simply through mixing with other haplogroups]".
ReplyDeleteI don't recall your offering any explanation as to how it might occurr.
"The main takeaway for you is that current mtDNA haplotypes were built in pre-ancient DNA times. Consequently, they miscode a number of ancestral retentions ascertained in Denisova (with or without Neandertals) as innovations".
Reaaly? What evidence do you base that statement on? I was certainly under the impression Denisova, Neanderthal and chimpanzee mt-DNA lines fit completely comprehendible patterns. What problems do you see?
"Plus they are full of reverse mutations and homoplasies. In a word, they need to be revisited at a fundamental level".
I agree with the first part but a great deal of research and re-arranging have mostly accounted for those factors.
"admixture can shift the root of a tree pretty dramatically because it brings a bunch of ancient sequences into a more recent population thus making it look older than it actually is".
Admixture cannot 'shift the root of a tree' at all. I agree if you believe all the haplogroups in a particular population appeared in theat population at the same time you will face age calculation problems, but such problems are easily avoided. The haploid genes themselves do not change with admixture in any way, shape or form.
"Mal'ta has other components in addition to the Amerindian one but it didn't provide Amerindians with a special Malta component".
Now we're getting somewhere. Yes, Mal'ta 'has other components in addition to the Amerindian one', but a population very similar to that the Mal'ta boy belonged to provided the Amerindians with the proportion of their genes shared with non-East Asian populations from Eurasia.
"the best way to think about the data at hand is that Mal'ta is an ancient East Asian population formed mostly by admixture from West Eurasia and the southern areas of the New World"
How could it possibly be admixture only with 'the southern areas of the New World'? What happened to the north American population it must have passed along the route?
"A more recent East Asian population, which we associate with the Mongoloid phenotype (EDAR, shovel-shaped incisors, etc.) is likely product of admixture between the already admixed ancient east Asian population and a new wave of northern Amerindians (relatives of Na-Dene and Eskimo-Aleuts) that brought the specific Mongoloid phenotype to East Asia after the retreat of the glacier around 12,000 years ago".
Isn't it far more likely that movement was in the other direction? That Na-Dene and Eskimo-Aleuts represent an increased level of Mongoloid admixture as compared to the bulk of the Amerindian population.
"Obviously we cannot say that the Mal'ta individual is from modern populations (for instance, 33% Tajik, 66% Udmurt), but can't we possibly see that the various admixture components (Gedrosia, Uralic, Amerind, etc.) are much older".
ReplyDeleteYes, except that, as Tobus said, 'Because that population wasn't well and alive in America at the time of Mal'ta'. So the component Mal'ta and Amerindians have in common must have come from Eurasia, not from America. Much the same may be true of the other populations you mention:
"Why would an individual show these populations, if they were not descended from these groups?"
An individual would show those populations if the populations were descended, at least in part, from the population the individual is a memeber of. Each of the other populations would likely be admixed with different sets of other populations.
" I don't think it's unreasonable to say that these things could be very old, near the East and West Eurasian split".
The pattern of haplogroup distribution shows the East and West Eurasian split is extremely ancient. We basically have two different sets of haplogroups, each having moved around opposite ends of the Himalayas. What is surprising about the current paper is that the Mal'ta boy completely lacks any East eurasian element. This strongly suggests the East Asians had not moved north of Northern China/Inner Mongolia at the time of the Mal'ta boy's existence whereas their haplogroups and aDNA later came to dominate the region.
"could we be seeing someone who is an offspring of a majority Gedrosian parent and a majority Uralic parent?"
Like the Native Americans the Uralic population were probably yet to form at the time. Certainly so in terms of their present haplogroups.
Although these maps are quite old now subsequent research has not really altered the distributions shown:
http://www.scs.illinois.edu/~mcdonald/WorldHaplogroupsMaps.pdf
If you have not seen this paper you will find it very interesting:
http://www.plosone.org/article/info:doi/10.1371/journal.pone.0071390
And this one sums up Y-chromosome phylogeny:
http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0049170
@TerryT
ReplyDelete"Admixture cannot 'shift the root of a tree' at all."
A good paper to read is The Impact of Founder Effects, Gene Flow, and European Admixture on Native American Genetic Diversity, by Keith Hunley and Meghan Healy. They show with further references that admixed populations tend to end up closer to the root of phylogenetic trees. So, purer populations may be less diverse than admixed populations, hence more downstream in phylogenetic trees. So, a phylogenetic tree may be a depiction of the extent of admixture, not the degree of descent.
"The haploid genes themselves do not change with admixture in any way, shape or form. "
Why not? If you have an ancient population and a new one comes in and completely absorbs it, the new population will end up looking older than it used to be, prior to the admixture event. People have no problems entertaining an idea that, say, Y-DNA A00 is the product of admixture with archaic hominins in Africa, or hg U5 in modern Europe is the product of admixture between agriculturalists and foragers.
"Reaaly? What evidence do you base that statement on?"
E.g., mtDNA L3, which is a clade that's found all over Africa and all over Eurasia and America is currently defined by a combination of bases at sites 769 1018 and 16311. But L3 agrees with Denisova at 769 (all African-specific mtDNA lineages agree with Neandertals), is uniquely derived at 1018 (all African specific mtDNA lineages agree with Denisova and Neandertals) and 16311 is hypervariable in modern humans with many actual L3 sequences classified as downstream from the L3 node (e.g., M1, M2a2, M4''67, etc.) carrying the ancestral site (C) and some actual African-specific sequences (L2, L5c2) carrying a derived site (T).
PhyloTree writes A769G, which means that they think A is ancestral, but it's G that's likely ancestral because it's found on Denisova. And L3 clusters with Denisova at this site, while L1'2'4'5'6 with all of the available Neandertal sequences.
That's the actual sequence reality at a key African-specific vs. general human mtDNA nexus. What we would ideally like to see, if the phylogeny is robust, are several uniquely derived bases compared to the Denisova outgroup in addition to a set of uniquely derived bases present in the L0'1'2'4'5'6 clade. But we don't have it. And the L3 situation is just one example. All major nodes up and down the tree have this problem.
"How could it possibly be admixture only with 'the southern areas of the New World'? What happened to the north American population it must have passed along the route? "
I meant with ancient New World populations whose best modern examples are located further down south. The north was involved in a later admixture event.
"Isn't it far more likely that movement was in the other direction? That Na-Dene and Eskimo-Aleuts represent an increased level of Mongoloid admixture as compared to the bulk of the Amerindian population."
EDAR and shovel-shaped incisors are arguably stronger expressed in America than in Asia, the earliest skulls with generalized Mongoloid morphology are in America, the Mongoloid phenotype is intrusive in Asia, Clovis-type points show a consistent pattern of moving northward across the relevant Paleoindian sites, a number of haploid lineages such as A2a are found back-migrated to Siberia, while such major Asian haplogroups as Y-DNA N and O are not found in America, the Tianyuan DNA is closer to Karitiana, etc. I don't know what you are basing your interpretation on. Mine is based on data.
"Yes, except that, as Tobus said, 'Because that population wasn't well and alive in America at the time of Mal'ta'. "
Well, Tobus happens to be wrong. See above.
@German:
ReplyDeleteSo you postulate the extinction of a proto-Amerindian population in Siberia and the lack of antiquity of the Amerindian population in America? Well, this is clearly biased and self-serving
Call it whatever you want, it's in line with all the evidence and is the scientific consensus among the experts in the field.
Mal'ta proves that Amerindians were around at 25,000 YBP
No it doesn't! I suggest you apply your "biased and self-serving" criticism to your own theories.
See the latest find at 30,000 YBP in Uruguay
This "latest find" is from 1997, it's only the confirmation of the dates that is recent. This is probably the best evidence to date for early humans in America but the authors themselves acknowledge that it's not definitive. As I've said before even *if* humans were in America pre-15kya, the genetic evidence tells us they weren't related to present day Amerindians, and so aren't the source of the "Amerindian" component in Mal'ta.
The first one is simplistic and incomplete as Mal'ta and West Eurasians are closer to Amerindians than they are to East Asians, which dramatically changes the meaning of your (factually correct) claim A
You seem to be agreeing that Amerindians are closer to East Asians than Amerindians are to Mal'ta. This means your original statement "modern Eurasian populations are less Amerindian than Mal'ta" is incorrect - some modern Eurasian populations (eg modern East Asians) are "more Amerindian" (to use your backwards terminology) than Mal'ta.
If East Asians are closer to Amerindians than they are to West Eurasians and West Eurasians are closer to Amerindians than they are to East Asians, it means Amerindians likely predated and spawned at least of these populations.
This is pure speculation. There are lots of other possibilities and the one that best fits the archaeological and genetic record is that Amerindians derive from an (ancient) East Asian population with a small amount of West Asian admixture. Perhaps it's time for another "biased and self-serving" assessment?
Your claim A was incomplete and claim B factually wrong
No, *your* initial claim was incomplete and my claim A corrects it - Amerindians are closer to East Asians, a fact you either weren't aware of or "forgot" to mention. You haven't presented any facts that contradict my claim B, only a series of personal speculations of a "likely" alternative. "Factually wrong" doesn't mean "different to what German made up".
@terryt
ReplyDeleteThank you, for the information. Lots of great links. I do have a few more questions about a few things. Anyone, feel free to chime in. I personally don't subscribe to the out-of-America hypothesis, for East Asians. My take on it is, that an ancestral population to American Indians, admixed into the Mal'ta population. Now, here is where I have questions. So, this paper says they picked up roughly the same amount of Amerindian as the MDLP k27 run. Now if that is correct, how can we say that the Amerind/Ancient Siberian is real, but not the Gedrosia, East European and Uralic. If this is really just a signature of people who Mal'ta admixed into, then why is it not including more West Eurasian groups besides East European Baltic? Why wouldn't it show more into other West Eurasian people, instead of more in the Papuan and Bantu categories? If we count the Amerind/Siberian as real, can we not call the Gedrosian and Uralic real? Do we really know when or how these populations started. If West and East Eurasians split roughly 40kya, isn't this plenty of time for separate components to begin to form? I understand your take on haplogroups; however, I am not so sure that haplogroups explained populations terribly much more than they do today. For instance, an R1a in India is not admixed like an R1a Pole. Someone who is R1b in Central Asia, is not like an R1b in Ireland. Perhaps our timeclock on all haplogroups is way off. Sorry, if my questions repeat, I am just trying to understand if stuff is really admixed from Mal'ta, why not have him score in my areas on a k27? Perhaps, I should save some questions until we have more remains. Thanks, in advance for your input.
"A good paper to read is The Impact of Founder Effects, Gene Flow, and European Admixture on Native American Genetic Diversity, by Keith Hunley and Meghan Healy. They show with further references that admixed populations tend to end up closer to the root of phylogenetic trees".
ReplyDeleteExcuse me! I've just read the abstract (can't get the whole paper at present) and it says:
"Our strategy is to test the fit of a serial founder effects process to the pattern of neutral autosomal genetic variation and to examine the contribution of gene flow and European admixture to departures from fit. The genetic data consist of 678 autosomal microsatellite loci assayed by Wang and colleagues in 530 individuals in 29 widely distributed Native American populations".
Note: 'autosomal genetic variation' and 'autosomal microsatellite loci'. No mention of haploid genes. Anyway we had a discussion on this paper at:
http://dienekes.blogspot.co.nz/2011/09/latent-admixture-causes-spurious-serial.html
"If you have an ancient population and a new one comes in and completely absorbs it, the new population will end up looking older than it used to be, prior to the admixture event".
Only in its autosomal genes. Any haploid genes within the admixed populations will remain, although some may be lost through drift. But the haploid genes themselves cannot possibly mix with each other. They are transmitted virtually unchanged from mother to child in the case of mt-DNA and from father to son in the case of Y-DNA. No mixing.
"People have no problems entertaining an idea that, say, Y-DNA A00 is the product of admixture with archaic hominins in Africa"
That is because the calculated age of A00 is older than th epresumed age of Homo sapiens. The haplogroup has changed since it diverged from haplogroups today found in all other humans but that change has in no way been influenced by any admixture.
"or hg U5 in modern Europe is the product of admixture between agriculturalists and foragers".
Haplogroup U's 'survival' in Europe is 'the product of admixture between agriculturalists and foragers', not the haplogroup itself.
"mtDNA L3, which is a clade that's found all over Africa and all over Eurasia and America is currently defined by a combination of bases at sites 769 1018 and 16311. But L3 agrees with Denisova at 769 (all African-specific mtDNA lineages agree with Neandertals), is uniquely derived at 1018 (all African specific mtDNA lineages agree with Denisova and Neandertals) and 16311 is hypervariable in modern humans"
You've hit the nail on the head with the last comment. Back mutation seems to be the answer to your problem. I'll leave others more expert in the field to cover the individual mutations you mention.
"Mongoloid phenotype is intrusive in Asia"
I agree it is intrusive north of Inner Mongolia, as the Mal'ta boy's DNA suggests.
"a number of haploid lineages such as A2a are found back-migrated to Siberia, while such major Asian haplogroups as Y-DNA N and O are not found in America"
I'm quite prepared to accept some A2a has entered northeastern Siberia from America, or more likely Beringia, but certainly didn't move very far into Eurasia. But N and O's absence from America surely indicates their presence in Siberia is the result of relatively recent northward movement from somewhere to the south.
"the Tianyuan DNA is closer to Karitiana"
Yes. Thus demonstrating the presence of an East Asian element, not present in the Mal'ta population, in Native Americans.
" I don't know what you are basing your interpretation on. Mine is based on data".
Your interpretation requires dismissing much data and considerably manipulating other data, indicating you have decided in advance what you want the data to reveal.
German, so, do you think that Amerinds had back-migrated to China already 40 kya, which is the age of the Tianyuan man?
ReplyDeleteChad Rohlfsen,
ReplyDeleteObviously we cannot say that the Mal'ta individual is from modern populations (for instance, 33% Tajik, 66% Udmurt), but can't we possibly see that the various admixture components (Gedrosia, Uralic, Amerind, etc.) are much older. Why would an individual show these populations, if they were not descended from these groups?
The genes in those populations are ancient, but the union of ancestrally diverse genes they represent are relatively modern (post MA-1). The problem is that there simply are no European-like Siberians who lack East Asian admixture, and no ANI/Kalash/Gedrosia-like populations that lack ancestral South Indian admixture any longer, so what we see is an approximation in oracle assignments based on a balance of two things: overall ancestral compatibility and degree of relatedness.
What I mean by that is, if we ignore the South Asian element for simplification, MA-1 is likely most strongly related to Amerindians. However, because modern Amerindians have significant (majority) ancestry from a population MA-1 has no relation to (i.e. East Asians), he isn't shown to be Amerindian in his oracle scoring, despite their abundant representation in MDLP dataset. On the other hand, his overall ancestry (discounting again the South Asian element) is likely most compatible with NE Europeans, maybe Lithuanians, but he's also fairly removed geographically and genetically from them compared to, say, Udmurts.
So what the oracle does in the absence of a perfect representative is to find a happy medium: Udmurts have the happiest ratio of strength of relatedness to unrelated ancestry (they're about 30% East Asian). But this certainly doesn't mean that Udmurt-like populations existed back then or that the East Asian in modern Udmurts was present in MA-1.
Think of it like this: if we discovered the genome of an ancient North African, and he lacked SSA admixture, we still shouldn't be surprised if his oracle scoring identified him as most resembling modern Berber populations, despite the latter having significant SSA admixture.
If these separations had not occurred, why would they appear in say a k27 run? I don't think it's unreasonable to say that these things could be very old, near the East and West Eurasian split. If I am wrong in my assumption, can someone please provide references to their answers (preferably peer-reviewed if possible).
A lot of what I mention above can be inferred from the supplementary information. Udmurts are unfortunately absent in the dataset used, so what I said can't be verified directly, but here are a few points:
SI 13 shows the Udmurt-like Mari receiving additional admixture from a population basal to (East Asian-admixed) modern Karitiana that the other W. Eurasian populations, including MA-1, lack. This is consistent with my point about the East Asian connection of modern Uralic populations being additional to the essential relationship between them and MA-1.
SI 21 is useful to contrast with the PCA Dienekes' article shows. It suggests that populations with high sharing of drift with MA-1 (i.e. as shown in SI 21) are pulled away from MA-1 proportionate to their East Asian ancestry, suggesting there's a balance between degree of relatedness and degree of unrelated admixture that needs to be factored in when interpreting MA-1's relationship to modern populations.
Perhaps with more specimens from the area and time-frame we will know more. The Gedrosian percentage has me wondering; could we be seeing someone who is an offspring of a majority Gedrosian parent and a majority Uralic parent?
ReplyDeleteIt's unlikely. He may be ancestral to the majority elements in Uralics and Gedrosians. He may be from a cousin population of the ancestors of the two groups. He may also be an admixture of two populations each independently related to one of those two populations. But, if so, it's unlikely he was bang in the midst of an admixture event. There's an interpretive bias toward seeing ancient DNA in extremely statistically unlikely ways that should be checked. He was most likely of the same admixture proportions as his mother and father, and his grandparents and great grandparents, given the likely isolation of the population. If admixture occurred, the chances of it being MA-1's generation specifically that received the admixture are slim.
Typos corrected:
ReplyDelete"but the union of ancestrally diverse genes they represent is relatively modern"
"MDLP's dataset"
Chad,
Now if that is correct, how can we say that the Amerind/Ancient Siberian is real, but not the Gedrosia, East European and Uralic. If this is really just a signature of people who Mal'ta admixed into, then why is it not including more West Eurasian groups besides East European Baltic?
Mal'ta folk likely weren't ancestral to many/any West Eurasian populations, but were just an easternmost cousin population, one most related (within W. Eurasia) to NE Europeans, ancestral north Indians and NW Europeans, but fundamentally related to all West Eurasians. This is seen (by inference) in figure SI 25 in the supplementary information.
ADMIXTURE analyses of aDNA are a bit of a minefield, since they're usually a medley of relationships to other populations, admixture from other populations, and admixture to other populations.
Why wouldn't it show more into other West Eurasian people, instead of more in the Papuan and Bantu categories?
Some ADMIXTURE analyses show 'Amerindian' admixture in all European and central/South Asian populations, and these percentages correspond very closely with the same populations' similarity to MA-1, so the connection isn't exclusively limited to Baltic and Eastern Europeans, even in ADMIXTURE analyses.
Compare k=4 here: https://docs.google.com/spreadsheet/ccc?key=0ArJDEoCgzRKedGR2ZWRoQ0VaWTc0dlV1cHh4ZUNJRUE#gid=1
with figure SI 21.
Why MA-1 doesn't score a percentage in all modern Western Eurasian categories at k=27 is probably the same reason I wouldn't score a percentage in all W. Eurasian categories: because I'm better represented by some W. Eurasian categories than by others, so much so that those categories leave little left over for, say, NW African, even if I'm more related to NW Africans (through common W. Eurasian-ness) than to East Asians.
Hamar, I have had difficulties in understanding the Figure SI 25. Does the plot A mean that Native Americans are all more related to West Eurasians than to Tianyuan and the biggest affinity is between Native Americans and Orcadians and the smallest between Native Americans and Basques (this same affinity is clearly visible also in the analysis you refer to). Does the plot B mean that East Asians are placed in between West Eurasians and Tianyuan, but East Asians are closer to Tianyuan than to Italians and to Russians while they are closer to Adyge, Burusho and Pathan than to Tianyuan? I cannot see any reference to Mal’ta here. In my opinion, the plot A argues against a big East Asian admixture of Amerinds, but the plot B argues for a western (ANI) admixture of modern East Asians compared to Tianyuan.
ReplyDeleteIn the admixture analysis you refer to, Han have 1% of Amerindian while even Bantus score percentages between 0.3-0.9. Lithuanians score even 9.2% of Amerindian. Oroquens score 13.2% of Amerindian, and, in fact, they harbour Native American mtDNA C1a with a frequency of even 11.5%.
I think that there has been movement of people from Asia to Europe and a part of Mal’ta’s West Asian ancestry could be genes that migrated from Asia to Europe.
Here http://www.biomedcentral.com/content/pdf/1471-2148-13-127.pdf you can find another admixture analysis of various Eurasian populations, Figure 6. At K=3 level, Burusho share 1/3 of their ancestry with East Asians and a tiny percentage with Native Americans. On Eurogenes Blogspot Ebizur said that “Approximately 25% of Burusho mtDNA belongs to macrohaplogroup M, but hardly any belongs to typical Northeast Asian/American subclades of M (only 1/44 in haplogroup C in the Hunza Burusho sample of Quintana-Murci et al. 2004, versus 10/44 in haplogroup M(xC, D, G, Z))”.
I still do not understand how Native Americans should have admixed heavily with an East Asian population while, at the same time, they are clearly closer to West Eurasians than East Asians (Figure SI 25). Moreover, in this other admixture analysis where Africans are absent, Native South Americans on no level show any affinity with East Asians.
Hamar, I have had difficulties in understanding the Figure SI 25. Does the plot A mean that Native Americans are all more related to West Eurasians than to Tianyuan and the biggest affinity is between Native Americans and Orcadians and the smallest between Native Americans and Basques (this same affinity is clearly visible also in the analysis you refer to).
ReplyDeleteIf you turn both plots 90 degrees clockwise and imagine the word 'Americans' where it says -0.10 and 'Tianyuan' where it says 0.10, it should make more sense. Plot A means that when West Eurasian populations are forced to 'choose' between Tianyuan and Americans based on genetic similarity, all choose Americans. This in itself suggests a special connection between Americans and W. Eurasians [at least largely] to the exclusion of a representative of the wider East Eurasian population, and this can either suggest flow from W. Eurasians to Americans, or flow from Americans to W. Eurasians (discounting the possibility of a more recent divergence between W. Eurasians and Americans than between W. Eurasians and other Asians, dealt with in part in plot B and in the remainder of the paper).
But if the flow was from Americans to W. Eurasians, then W. Eurasians should also have inherited some of the secondary within-East-Eurasian relations that Americans show. So if Americans are substantially more related to Han than to Papuans (which they are), then if W. Eurasians are significantly part American, they should also be significantly more related to Han than to Papuans, but they aren't (the figure for this is only in the main paper). So the figure is basically showing that W. Eurasians are preferentially related to Americans vs. Tianyuan, and some additional figures show that W. Eurasians are not preferentially related to other E. Eurasian populations, even when Americans are, suggesting the direction of flow is from W. Eurasia to America.
Image B does a similar thing: it forces W. Eurasians to choose between Tianyuan and modern Asians, but there's little to no pattern in that relationship, suggesting that, among other things, internal East Eurasian factors seem not to be responsible for the pattern in plot A.
Does the plot B mean that East Asians are placed in between West Eurasians and Tianyuan, but East Asians are closer to Tianyuan than to Italians and to Russians while they are closer to Adyge, Burusho and Pathan than to Tianyuan? I cannot see any reference to Mal’ta here.
What I meant was that, unless there were two or more sources for the West Eurasian signal in Amerindians, then Mal'ta (or, more, realistically, an unknown population he's a proxy for) is the source of the relationship in plot A. Otherwise, we'd need an injection into the Americas of a population related to Europeans and S/C Asians on the one hand and then another one related to SW Asians and North Africans on the other to explain plot A. I think the best interpretation is that Mal'ta was primarily related to Northern Europeans and S/C Asians, but secondarily related to all West Eurasians, but not necessarily ancestral to any of them.
In the admixture analysis you refer to, Han have 1% of Amerindian while even Bantus score percentages between 0.3-0.9. Lithuanians score even 9.2% of Amerindian. Oroquens score 13.2% of Amerindian, and, in fact, they harbour Native American mtDNA C1a with a frequency of even 11.5%.
ReplyDeleteKeep in mind that in the analysis, Amerindian populations largely score 100% Amerindian, so that component encompasses both the East and West Eurasian elements of their ancestry. But because ADMIXTURE considers Amerindians one single population, a certain degree of high-level noise is projected into W. Eurasians on the one hand and East Asians on the other, because their similarity to portions of modern American ancestry is interpreted as admixture, since Amerindians aren't considered mixed. Therefore, 'Amerindian' percentages can be scored and shared by populations with no genuine genetic overlap (such as Han and Basques). If you look at k=3 you'll see that some populations with Amerindian at k=4 are 100% European or 100% Asian (such as Basques and Sardinians, Han and Japanese). This is consistent only if 'Amerindian' admixture doesn't necessarily equal East Asian admixture, but also can mean that in certain instances. In populations where the 'Amerindian' signal at k=4 seems independent of East Asian at k=3, the Amerindian score mirrors the relationship with MA-1.
As for sub-Saharan Africans, there's quite a bit of European and Asian noise for the same populations. Some of it may even be real, though. The EDAR gene appears in one Yoruban here, so very low-level geneflow seems to have occurred between East Asia and Africa (obviously indirectly):
http://alfred.med.yale.edu/alfred/mvograph.asp?siteuid=SI663326A
Here http://www.biomedcentral.com/content/pdf/1471-2148-13-127.pdf you can find another admixture analysis of various Eurasian populations, Figure 6. At K=3 level, Burusho share 1/3 of their ancestry with East Asians and a tiny percentage with Native Americans.
The majority of sharing between Burusho and East Asians is via Burusho's ancestral south Indian heritage. Ancestral south Indians are East Eurasian, but not East Asian. The Amerindian component presumably shows for the same reason it shows in Europeans. Burusho also have a minor amount of real East Asian ancestry. They have 6% EDAR at the same link as above:
http://alfred.med.yale.edu/alfred/mvograph.asp?siteuid=SI663326A
I still do not understand how Native Americans should have admixed heavily with an East Asian population while, at the same time, they are clearly closer to West Eurasians than East Asians (Figure SI 25). Moreover, in this other admixture analysis where Africans are absent, Native South Americans on no level show any affinity with East Asians.
Figure SI 24 shows the relationship of Amerindian populations to Han vs. MA-1. Most are more related to Han. Of course, we'd need to find as good an ancient proxy for the direct contributor of East Asian genes to Amerindians as Mal'ta is for the contributor of W. Eurasian genes before we based admixture proportion estimates from that figure alone.
@Kristina: I still do not understand how Native Americans should have admixed heavily with an East Asian population while, at the same time, they are clearly closer to West Eurasians than East Asians (Figure SI 25).
ReplyDeleteI think Hamar's comments are really good, but I'd like to stress that Figure SI 25 is primarily looking at those SNP's that are *different* between Americans and East Asians. The D-statistic works by counting ABAB vs ABBA so it's implicit that any allele shared between the two populations pairs on each side are ignored... so in D(Africa, X, Tianyuan, America) all SNPs common to Tianyuan and America (ABBB or BABB) are excluded from the count. Likewise the D(..., Tianyuan, East Asian) chart only uses alleles that East Asians have and Tianyuan doesn't. So the two charts together are only comparing affinity to the post-Tianyuan divergence part of Americans and East Asians, not to the populations as a whole. There could be 90% similarity between the two but the charts would still show other populations "closer" to one or the other because it's designed to compare only the 10% that is different. In short S25 doesn't show Native American "are clearly closer to West Eurasians than East Asians" because it has deliberately excludes most of the American/East Asian shared alleles from the study. What it's saying is that *where Americans and East Asians are different* Americans are closer to West Eurasians... this is not the same as saying that they are closer across the board.
I'll also note that Americans and East Asians being "heavily admixed" is not the way I'd put it. It seems they both share the same Tianyuan-like ancestors in East Asia, so their affinity has primarily come from the divergence of a single ancient population, not admixture between two existing populations.
@ Kristiina. I've just seen your comment at Maju's blog (where I have been banned) suggesting both mt-DNAs M8'C and M80'D are Central Asian in origin. That is most unlikely. Although C is virtually absent in SE Asia M8 is especially common there, especially in Vietnam and Laos. As far as I'm aware M80 is confined to Palawan, today politically part of the Philippines but geographically connected to Borneo. D is widespread through SE and East Asia. The distribution of M haplogroups as a whole strongly suggests an original expansion from the hill country between northeast India, burma and South China. I also dissagree strongly with Maju that M entered Australia before either N or R did.
ReplyDelete@ Chad Rohlfsen:
"My take on it is, that an ancestral population to American Indians, admixed into the Mal'ta population".
In a way. I don't think 'American Indians, admixed into the Mal'ta population', by the time they entered America they had become an admixed population of Mal'ta and East Asian populations with East Asians making up the majority. Later arrivals such as Na-Dene and Inuit were more heavily East Asian.
"If we count the Amerind/Siberian as real, can we not call the Gedrosian and Uralic real? Do we really know when or how these populations started"
No, we don't. But we are getting closer. Hamar Fox explains the remainder of this aspect far better than I could.
"I am not so sure that haplogroups explained populations terribly much more than they do today".
Correct. And that goes even for species, as the paper on the 400,000 year old specimen from Spain shows. However haplogroups can give us a very good idea of population expansion. When a set of genes expands they would often be carried by either or both mt- and Y-DNA although haplogroups often expand further than do diploid genes. Languages expand even further.
"Perhaps our timeclock on all haplogroups is way off".
I don't have a time on haplogroup diversification at all. In fact I fail to see why survival of mutations would be regular in any way. However what we can discern from haplogroup phylogeny is the order of appearance of particular haplogroups. That is the bit German has so much trouble agreeing with.
@ Hamar Fox:
"Of course, we'd need to find as good an ancient proxy for the direct contributor of East Asian genes to Amerindians as Mal'ta is for the contributor of W. Eurasian genes before we based admixture proportion estimates from that figure alone".
Yes. And I think we both have a good idea of where we could find the main source of the modern East Asian element: slightly east of Tianyuan, in the higher altitude regions. But useful fossils may not exist.
Moreover, in this other admixture analysis where Africans are absent, Native South Americans on no level show any affinity with East Asians.
ReplyDeleteTo see their affinity you must look at K=2, which is unfortunately not shown in that paper. The "East Eurasian" and "Amerindian" components at higher levels are much closer to each other than any of them is to any one of the "West Eurasian" components.
Ancestral south Indians are East Eurasian, but not East Asian.
No, the Ancestral South Indians (=ASI) are intermediate between West and East Eurasians, probably slightly closer to East Eurasians.
The Amerindian component presumably shows for the same reason it shows in Europeans.
In that ADMIXTURE analysis the French, North Italians, Tuscans, the Balochi, the Brahui and the Makrani have no "Amerindian" components or at noise level at most, so the small "Amerindian" components of the Burusho, which are bigger than those of Pathans and the Sindhi, must mostly or wholly be part of the Mongoloid admixture of the Burusho.
terryt wrote,
ReplyDelete"Yes. And I think we both have a good idea of where we could find the main source of the modern East Asian element: slightly east of Tianyuan, in the higher altitude regions. But useful fossils may not exist."
What "higher altitude regions" that exist "slightly east of Tianyuan" are you imagining? Some part of Korea or Japan? If not, then your comment does not make any sense. Tianyuan is a part of the Zhoukoudian complex (from which the famous "Peking Man" fossil of Homo erectus has been unearthed) on the outskirts of Beijing.
In light of Tobus' clarification, I should say that SI 24 doesn't show whether the Amerindian populations included are closer to MA-1 or Han, but whether they're closer to MA-1 or Han relative to Karitiana. But what I said still applies, I think, to a PCA-based analysis of ancestral proportions.
ReplyDeleteOnur,
No, the Ancestral South Indians (=ASI) are intermediate between West and East Eurasians, probably slightly closer to East Eurasians.
Possibly so, but the matter's obscured by the fact that 'South Asian' components are pretty much always composites of ANI and ASI, which in itself could explain the South Asian component's intermediacy between West and East; and the ANI/ASI balance of S. Asian components also shifts drastically not only from calculator to calculator, but from k-level to k-level within the same calculator-series (such as Dodecad world 3-13), so that makes it even harder to pin down and is responsible for inconsistent proportion estimations of S. Asian in various populations, and also for the ever-shifting fst values of that component.
Exclusively South Indian components tend to appear only at high k-levels, and could well also be mixed. At any rate, the Burusho's East Eurasian affiliations are still mostly by way of ASI admixture.
In that ADMIXTURE analysis the French, North Italians, Tuscans, the Balochi, the Brahui and the Makrani have no "Amerindian" components or at noise level at most, so the small "Amerindian" components of the Burusho, which are bigger than those of Pathans and the Sindhi, must mostly or wholly be part of the Mongoloid admixture of the Burusho.
Also possible, but within that calculator, based on the poulations included, French, N. Italians and Tuscans constitute the W. Eurasian pinnacle, so if ADMIXTURE detects more of an Amerindian signature in Burusho than in the archetypal populations, it will say that Burusho have X amount of Amerindian, where X is how much more Burusho have than French, N. Italians and Tuscans -- provided that, for whatever reason, the algorithm doesn't also calculate that the 'pinnacle' populations are also admixed (which, as we know, sometimes happens and sometimes doesn't).
The same is true within the East Asian populations; e.g. Han vs. Hezhen.
But I definitely agree with you that it wouldn't be surprising if what should be behind/in place of the brown in many populations turned out to be yellow and not blue, because I've seen ADMIXTURE get confused by its own assumptions sometimes.
Thank you for very good explanations! Now I understand the plots much better.
ReplyDeleteTerry, I did not propose that mtDNA M80'D has a Central Asian origin. Instead, I suggested that mtDNAs M8C and A might have a Central Asian origin. The oldest branch of M80'D seems to be D5, and it is mostly a Chinese and Japanese/Korean lineage. According to the Chinese mtDNA N paper (Chen Zhiyong) and "A Mitochondrial Revelation of Early Human Migrations to the Tibetan Plateau Before and After the Last Glacial Maximum”, 2010", the northermost branches seem to be the oldest (American A2 and Tibetan A10). As for mtDNA C, it is well known that it is found mainly in Altai, America, Central Asia/Siberia and India, but it is present also in Iran, Caucasus and Europe, and detected in ancient sites in the West. Of course, this debate has been going on since Oppenheimer's out of Eden.
It is true that I am eager to see multiple different routes and not only a few highways for the human migrations.
"What 'higher altitude regions' that exist "slightly east of Tianyuan" are you imagining?"
ReplyDeleteSilly me. I got my geography reversed. Should have said, 'slightly west of Tianyuan'. Plenty of higher altitude regions there.
"Tianyuan is a part of the Zhoukoudian complex (from which the famous 'Peking Man' fossil of Homo erectus has been unearthed) on the outskirts of Beijing".
Particularly interesting in this context as it has mt-DNA B, one of the American haplogroups originally from East Asia.
" I suggested that mtDNAs M8C and A might have a Central Asian origin".
ReplyDeleteI agree completely that A has 'a Central Asian origin'. In fact it is extremely interesting that there are no exclusively South Asian basal N haplogroups, in spite of many people wishing to believe N moved east through that region. I don't see an 'M8c' in Phylotree. Which branch of M8 do you mean?
"The oldest branch of M80'D seems to be D5, and it is mostly a Chinese and Japanese/Korean lineage".
D5 may be the oldest 'D' haplogroup but the branch leading to M80 must be older than any diversification of D itself. I would guess that both D and M8'CZ moved north from Yunnan through the hill country bordering Tibet rather than through 'China'. The heavily vegetated, and perhaps swampy, Chinese plain may have been virtually unoccupied until the spread of farming. On the other hand R-derived B may have moved north along the coast.
"It is true that I am eager to see multiple different routes and not only a few highways for the human migrations".
Of course. The alternative is what I refer to as the Garden of Eden syndrome.
Possibly so, but the matter's obscured by the fact that 'South Asian' components are pretty much always composites of ANI and ASI, which in itself could explain the South Asian component's intermediacy between West and East
ReplyDeleteI was referring only to ASI, not to the "South Asian" components. But I made a typo in my statement. I wrote "probably slightly closer to East Eurasians" for ASI. I should have written "probably significantly closer to East Eurasians" instead. But I think it is a mistake to define ASI in terms of West Eurasianness and East Eurasianness. It is a separate race with distinct genetic and phenotypic characteristics. Its "intermediacy" is only partial. Otherwise in Eurasian-wide PCAs it would fall on the West Eurasian-East Eurasian cline that makes up the 1st dimension of such PCAs, but it does not, and falls on a quite removed position forming the 2nd dimension.
At any rate, the Burusho's East Eurasian affiliations are still mostly by way of ASI admixture.
I did not object to that. In fact, I agree with that statement.
@Hamar Fox
ReplyDelete"But if the flow was from Americans to W. Eurasians, then W. Eurasians should also have inherited some of the secondary within-East-Eurasian relations that Americans show."
Not if East Asians are derived from Amerindians. Gene flow couldn't have been from Mal'ta to Amerindians as all of West Eurasians are closer to Amerindians than they are to East Asians. And Amerindians couldn't have derived from East Asians because there were no East Asians in Siberia at the time of Mal'ta when the Amerindian component was already present.
"@ Hamar Fox:
"Of course, we'd need to find as good an ancient proxy for the direct contributor of East Asian genes to Amerindians as Mal'ta is for the contributor of W. Eurasian genes before we based admixture proportion estimates from that figure alone". "
What Terry wrote makes it obvious that Hamar Fox is wrong: Mal'ta did not contribute a West Eurasian component to Amerindians: BLUE is not found in the ADMIXTURE results for America, Y-DNA R and mtDNA U are not found in America either.
@Kristiina
"German, so, do you think that Amerinds had back-migrated to China already 40 kya, which is the age of the Tianyuan man?"
Yes. Considering that Tianyuan man is closer to Karitiana than to any other population and considering that Karitiana is attested 40,000 years later than Tianyuan, we should expect ancient DNA from America to provide further evidence to support my point. Notably, Mal'ta is another ancient DNA sample and it confirms the greater proximity of ancient Eurasians to modern Amerindians than to modern Eurasians.
mtDNA hg B is likely a relic haplogroup later overrun by a host of others. It survived in its more or less pristine frequencies on both sides of the Circumpacific region.
@terryT
"However what we can discern from haplogroup phylogeny is the order of appearance of particular haplogroups. That is the bit German has so much trouble agreeing with."
I gave you a clear example why I think the order of mutations on the mtDNA phylogeny is problematic. There's nothing to agree with - the model has a lot of noise and without ancient DNA from Africa - which is nowhere near to be coming - it's just an ideal model.
Onur,
ReplyDeleteI was referring only to ASI, not to the "South Asian" components. But I made a typo in my statement. I wrote "probably slightly closer to East Eurasians" for ASI. I should have written "probably significantly closer to East Eurasians" instead.
With that amendment, I fully agree with you.
But I think it is a mistake to define ASI in terms of West Eurasianness and East Eurasianness. It is a separate race with distinct genetic and phenotypic characteristics. Its "intermediacy" is only partial. Otherwise in Eurasian-wide PCAs it would fall on the West Eurasian-East Eurasian cline that makes up the 1st dimension of such PCAs, but it does not, and falls on a quite removed position forming the 2nd dimension.
I don't contest this. My statement was merely that when ADMIXTURE boils populations down to the basics, then it generally places both ASI-admixed (to the general extent of their inferred ASI admixture), E. Asian and Australasian populations in the same broad category, and that was the explanation of the phenomenon relevant to Kristiina's point at the time.
German Dziebel,
Not if East Asians are derived from Amerindians. Gene flow couldn't have been from Mal'ta to Amerindians as all of West Eurasians are closer to Amerindians than they are to East Asians.
I know your interpretation of events, German. I don't quite understand your above point, though, since it hinges on the assumption of MA-1 not being related to all W. Eurasians (since if he was, it's obvious how his admixture into Amerindians would forge a special relationship between Amerindians and all W. Eurasians). But even in the ADMIXTURE analysis you put so much stock in, MA-1 shares components with all W. Eurasians until k8 (where the component in Mal'ta becomes N. European dark blue at the expense of the wider W. Eurasian light blue).
And Amerindians couldn't have derived from East Asians because there were no East Asians in Siberia at the time of Mal'ta when the Amerindian component was already present.
Above I said this, and it applies here:
"ADMIXTURE analyses of aDNA are a bit of a minefield, since they're usually a medley of relationships to other populations, admixture from other populations, and admixture to other populations."
In addition, that specific ADMIXTURE analysis identifies Amerindians as exclusively East Asian/Eurasian at k=3. Given the other methods of assessing Amerindian genetic makeup, this seems to be a mistake on ADMIXTURE's part. So because the program (wrongly) considers all Amerindian ancestry/relations to be East Eurasian, it misidentifies the portion of MA-1's genome that resembles Amerindians (which he contributed) as also being East Eurasian at k=3. If you were to enter MA-1 into calculators that recognised Amerindians to be admixed at k=3, I imagine MA-1 would appear much less than 1/3rd East Eurasian.
What Terry wrote makes it obvious that Hamar Fox is wrong: Mal'ta did not contribute a West Eurasian component to Amerindians: BLUE is not found in the ADMIXTURE results for America, Y-DNA R and mtDNA U are not found in America either.
ReplyDeleteI'm not sure what you mean here. Terry's comment was in relation to my comment on D-statistics and the best references for West and East Eurasian contributive populations to Amerindians. It had nothing to do with ADMIXTURE.
If you're talking about the ADMIXTURE analysis Kristiina linked to, then I doubly don't get what you're talking about. If you want to take it at face value, that analysis shows that French, Tuscans and N. Italians have no Amerindian in them, eight East Asian populations have no Amerindian in them, and that four Amerindian populations have no European or East Asian in them, so how does it support your case that Amerindians are progenitors of all any more than mine? Again, at face value it says there's no fundamental relation or fundamental admixture between any of the three major components identified.
All it actually means, however, is that in the absence of SSA representatives, the three most distinct components are W. Eurasian, E. Eurasian (to the exclusion of Amerindians) and Amerindian, and it gives a value of 100% membership of those components to the populations the program determines best represent them.
Kalash also show as 100% Kalash when the component 'Kalash' appears. Does that mean they have no admixture in them whatsoever?
@German:Considering that Tianyuan man is closer to Karitiana than to any other population and considering that Karitiana is attested 40,000 years later than Tianyuan, we should expect ancient DNA from America to provide further evidence to support my point.
ReplyDeleteThe problem with this theory is that if the genetic line went Amerindian->Tianyuan(-like)->East Asian in Asia while Amerindians remained unadmixed in America, then modern East Asians should be closer to Tianyuan than modern Amerindians are. Even if EDAR and Mongoloid features are recent American developments as you say - East Asians would be Tianyuan/American hybrids and so should still appear closer to Tianyuan than "pure" Amerindians do.
Notably, Mal'ta is another ancient DNA sample and it confirms the greater proximity of ancient Eurasians to modern Amerindians than to modern Eurasians.
ADMIXTURE shows Mal'ta to be some 70% "modern Eurasian", so what you say appears to be incorrect - by that reckoning he has twice the proximity to modern Eurasians as he does to modern Amerindians. I suspect you meant "modern East Asians".
@Tobus
ReplyDelete"The problem with this theory is that if the genetic line went Amerindian->Tianyuan(-like)->East Asian in Asia while Amerindians remained unadmixed in America, then modern East Asians should be closer to Tianyuan than modern Amerindians are. "
No. And you know why:
"EDAR and Mongoloid features are recent American developments as you say," so there was a second migration out of the New World.
Your claim that "East Asians would be Tianyuan/American hybrids and so should still appear closer to Tianyuan than "pure" Amerindians do" doesn't seem to be right because:
If Tianyuan is the first wave out of America (the best surviving unadmixed examples being modern South Amerindians) and modern East Asians is the second one (the best surviving examples being Northern Amerindians), then Tianyuan will be closer to such populations as Karitiana, while modern East Asians to such populations as Na-Dene and Eskimo-Aleuts. This is exactly what we see between the data from Fu et al. and the data from Raghavan et al. East Asians will be closer to Tianyuan than Na-Dene and Eskimo-Aleuts because they absorbed a Tianyuan-like population as they expanded from the north into south China and beyond.
"ADMIXTURE shows Mal'ta to be some 70% "modern Eurasian", so what you say appears to be incorrect - by that reckoning he has twice the proximity to modern Eurasians as he does to modern Amerindians. I suspect you meant "modern East Asians"."
But Fig. 1C and Figure SI 22 show Amerindians having the largest amount of shared drift with Mal'ta out of all non-African populations. This makes me think that the Amerindian component in Mal'ta is older than the West Eurasian component, so that your 70% got accrued after the founding migration of Amerindians into the Old World.
@Hamar Fox
ReplyDeleteI was referring not to Kristiina's link but to your claim, which Terry repeated, that "we'd need to find as good an ancient proxy for the direct contributor of East Asian genes to Amerindians as Mal'ta is for the contributor of W. Eurasian genes."
But whether we take ADMIXTURE or Y-DNA or mtDNA results from Mal'ta, it's obvious that no West Eurasian genes were contributed to the New World. That's all.
"Kalash also show as 100% Kalash when the component 'Kalash' appears. Does that mean they have no admixture in them whatsoever?"
Populations such as Kalash or Hadza (see Henn 2011 or Tishkoff 2009), which tend to show less admixture than surrounding populations or no admixture at all, have indeed been unadmixed, due to their isolated, refugial geographic location, for a long time. Hadza tends to have lower intragroup allele diversity values than other Africans, but higher Fst and LD. Kalash likely, too, but I don't have the data at hand. Just like Amerindians on the global scale. Whatever ancient admixture these populations may have absorbed, there have been enough time for the allele frequencies to get equilibrated through random mating. Overall, the population genetic profile of Kalash and Hadza is indeed most similar to that of Amerindians.
German,
ReplyDeleteI was referring not to Kristiina's link but to your claim, which Terry repeated, that "we'd need to find as good an ancient proxy for the direct contributor of East Asian genes to Amerindians as Mal'ta is for the contributor of W. Eurasian genes."
Sorry, but where is the refutation and what is it that's supposedly being refuted? I honestly don't have a clue what you're talking about. If you think Terry refuted me by repeating what I said, isn't that just the same as saying I refuted myself? Why are the things you're talking about in reference to the quote not even remotely linked to anything mentioned or even alluded to in the quote? Argh, You're messing with my mind. I can't tell if anything's real anymore!
Populations such as Kalash or Hadza (see Henn 2011 or Tishkoff 2009), which tend to show less admixture than surrounding populations or no admixture at all, have indeed been unadmixed, due to their isolated, refugial geographic location, for a long time.
This is patently false. Kalash evidence the same admixture as surrounding populations up to the point that their own component emerges, which allows them to occupy it fully. This is an effect of isolation and the corresponding inbreeding: descent from a smaller set of common ancestors relative to other populations, shared unique drift, enhanced internal relatedness, a hastened homogenisation process etc. All that's needed really is a homogenous population (relative to others) with a large amount of shared genetic signature less intense or lacking in other populations, and eventually it will get its own component. All ADMIXTURE does is to prioritise some components over others.
Ancient admixture doesn't interfere with this process whatsoever, only recent admixture does, and only because it upsets the homogeneity until it levels out again. Indeed, most of the populations likely to get their own component on a 20-30 component calculator (Berber populations, Kalash, Saami etc.) are fundamentally admixtures of two or more highly divergent populations.
Hadza tends to have lower intragroup allele diversity values than other Africans, but higher Fst and LD. Kalash likely, too, but I don't have the data at hand. Just like Amerindians on the global scale. Whatever ancient admixture these populations may have absorbed, there have been enough time for the allele frequencies to get equilibrated through random mating. Overall, the population genetic profile of Kalash and Hadza is indeed most similar to that of Amerindians.
You seem to be making my case for me there, especially with this:
"Whatever ancient admixture these populations may have absorbed, there have been enough time for the allele frequencies to get equilibrated through random mating."
The admixture in Amerindians is absolutely equilibriated.
"mtDNA hg B is likely a relic haplogroup later overrun by a host of others. It survived in its more or less pristine frequencies on both sides of the Circumpacific region".
ReplyDeleteIts first arrival in the southwest Pacific is only some 4000 years ago. Hardly the signature of 'a relic haplogroup later overrun by a host of others'.
"Y-DNA R and mtDNA U are not found in America either".
I was sure everyone accepted by now that haploid DNA does not necessarily have a close correlation with diploid DNA.
"I gave you a clear example why I think the order of mutations on the mtDNA phylogeny is problematic".
I realise science is not a democracy but you seem to represent a minority of one. The phylogeny has undergone rigorous examination and although there remain some minor disagreements it appears well enough established for most scientists to accept.
"Gene flow couldn't have been from Mal'ta to Amerindians as all of West Eurasians are closer to Amerindians than they are to East Asians".
The second comment does not necessarily demand the first. If West Eurasians have no (or very little) East Eurasian genetic component while Amerindians have a significant amount we would also see that 'West Eurasians are closer to Amerindians than they are to East Asians'. And gene flow could very much have come into Amerindians from some Mal'ta-like population. That actually fits all the data far closer than the scenario you propose.
"Amerindians couldn't have derived from East Asians because there were no East Asians in Siberia at the time of Mal'ta when the Amerindian component was already present".
For a start we don't actually know that 'the Amerindian component was already present'. That is just your assumption, required for your theory to be correct. Secondly, although we know that 'there were no East Asians in Siberia at the time of Mal'ta' we do know they were present a little to the south. This explains why Amerindians have largely West Eurasian Y-DNA and mostly East Eurasian mt-DNA. They mixed.
@German:
ReplyDeleteIf Tianyuan is the first wave out of America (the best surviving unadmixed examples being modern South Amerindians) and modern East Asians is the second one (the best surviving examples being Northern Amerindians), then Tianyuan will be closer to such populations as Karitiana, while modern East Asians to such populations as Na-Dene and Eskimo-Aleuts
What!? An ancient American population splits into 2 lineages, one which goes into Asia and one that stays in America. Some time later the one that stayed in America also splits, with one population staying where it is and the other going to Asia and mixing with the earlier Asian lineage... and you think the population that stayed in America will be closer to the first Asian population than the one that has admixed with them since the initial divergence?!? I suspect you've forgotten to divulge some important aspect of your speculations, because it makes no sense the way it is.
East Asians will be closer to Tianyuan than Na-Dene and Eskimo-Aleuts because they absorbed a Tianyuan-like population as they expanded from the north into south China and beyond.
So you acknowledge the effect Tianyuan admixture in regard to East Asians vs Na-dene and Eskimo-Aleuts, but not in regard to East Asians vs Karitiana? What's special about Karitiana that they possess more genetic affinity with Tianyuan after over 40,000 years of isolation than other populations with the exact same shared ancestry but who also have more recent admixture with Tianyuan's descendants?
I can't see any way to make your theory work - if Amerindians are ancestors of both Tianyuan and East Asians then East Asians should be closer to Tianyuan than Amerindians are. Since they're not, it must be the other way round - Tianyuan is ancestral to both Amerindians and East Asians.
This makes me think that the Amerindian component in Mal'ta is older than the West Eurasian component
You are more than welcome to think that if you want, but please don't state that Mal'ta "confirms the greater proximity of ancient Eurasians to modern Amerindians than to modern Eurasians" when it clearly doesn't. It's the second time now that you've intentionally misrepresented the facts by attributing them to a larger population grouping than they actually apply to. A sound theory doesn't need subterfuge and misrepresentation to gain traction - if you have to resort to misleading statements of pseudo-facts to get your point across, perhaps it's time to reassess if the point is worth making.