Going over the 322 pages of thesis may take a while, but feel free to comment on it if you discover any interesting nuggets in the text. The following view of West/East Eurasian mtDNA surrounding the beginning of the Iron Age may be useful, and seems to parallel the results of a recent paper on Pazyryk mtDNA:
Of course, since the thesis was published we have new data from West Siberia/Ukraine that suggest that the penetration of east Eurasian lineages covered a great area to the west of the indicated region even prior to the Iron Age.
We can be fairly sure that "non-East Eurasian admixed" populations existed during the Bronze Age in three portions of the Eurasian landmass, separated by the Black and Caspian Seas: west of the Black Sea (Balkans/Central Europe); between Black and Caspian Seas (Caucasus) and east of the Caspian Sea (Kazakhstan and Turkmenistan). But how did these three regions contribute to the West Eurasian elements found on a west-east axis across Eurasia today? And, to what extent did the early east Eurasian elements that penetrated well into eastern Europe in the Neolithic-to-Bronze Age contribute to latter populations of the area vs. more recent expansions from the Altai and Central Asia during the Iron Age?
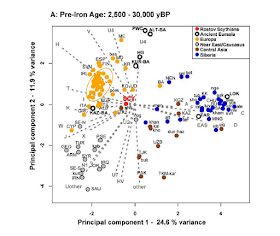
Here is a PCA of the pre-Iron Age individuals, compared with modern populations:
Both "Tarim" (TAR) and "Neolithic Lake Baikal" (LOK) appear well within east Eurasian variation. But, of the West Eurasian groups, Pitted Ware Complex (PWC), i.e., Neolithic hunter-gatherers from NE Europe and Bronze Age Altai (ALT-BA) appear clearly "northern Europeoid" across the 2nd PC, as do, to a lesser extent, C/N European Hunter-Gatherers (HG) and Kurgan burials from south Siberia (KUR-BA), but Bronze Age Kazakhstan (KAZ-BA) appear to be southern Europeoid, and, also, noticeably more "West Eurasian" than the others. Clearly, the West Eurasian elements were not homogeneous, with some of them (such as KAZ-BA) apparently derived from the southern Caucasoid zone -which largely did not experience east Eurasian admixture- and others from the northern Caucasoid zone that did.
The Rostov Scythian sample (in red) appears to belong to the southern Caucasoid zone (across PC2), but East Eurasian-shifted relative to modern Europeans and Bronze Age Kazakhstan.
Now, let's look at the Iron and post-Iron Age samples:
Egyin Gol (EG) from Mongolia and Sargat Siberians appear clearly as East Eurasians; Pazyryk Altai (ALT-IA), Iron Age Kazakhstan (KAZ-IA) and South Siberia Kurgan (KUR-IA) show decreasing East Eurasian influence; also notice the decidedly "southern" shift of the West Eurasian element among them.
This seems broadly consistent with the ideas of Molodin et al. about the gradual appearance (in their Siberian sample) of Caucasoid mtDNA types from the Neolithic to the Iron Age, with the early Neolithic U-dominated population finally receiving a full set of diverse West Eurasian lineages only during the Iron Age from the south.
It will certainly be very exciting when samples such as these can be tested for autosomal or Y-chromosome DNA, and I'm looking forward to the day when this can be done on a large scale.
Type: Thesis
Title: Mitochondrial DNA in ancient human populations of Europe.
Author: Dersarkissian, Clio Simone Irmgard
Issue Date: 2011
School/Discipline: School of Earth and Environmental Sciences
Abstract: The distribution of human genetic variability is the result of thousand years of human evolutionary and population history. Geographical variation in the nonrecombining maternally inherited mitochondrial DNA has been studied in a wide array of modern populations in order to reconstruct the migrations that have participated in the spread of our ancestors on the planet. However, population genetic processes (e.g., replacement, genetic drift) can significantly bias the reconstruction and timing of past migratory and demographic events inferred from the analysis of modern-day marker distributions. This can lead to erroneous interpretations of ancient human population history, a problem that potentially could be circumvented by the direct assessment of genetic diversity in ancient humans. Despite important methodological problems associated with contamination and post-mortem degradation of ancient DNA, mitochondrial data have been previously obtained for a few spatially and temporally diverse European populations. Mitochondrial data revealed additional levels of complexity in the population history of Europeans that had remained unknown from the study of modern populations. This justifies the relevance of broadening the sampling of ancient mitochondrial DNA in both time and space. This study aims at filling gaps in the knowledge of the genetic history of eastern Europeans and of European genetic outliers, the Saami and the Sardinians. This study presents a significant extension to the knowledge of past human mitochondrial diversity. Ancient remains temporally-sampled from three groups of European populations have been examined: north east Europeans (200 – 8,000 years before present; N = 76), Iron Age Scythians of the Rostov area, Russia (2,300 – 2,600 years before present; N = 16), Bronze Age individuals of central Sardinia, Italy (3,200 – 3,400 years before present; N = 16). The genetic characterisation of these populations principally relied on sequencing of the mitochondrial control region and typing of single nucleotide polymorphisms in the coding region. Changes in mitochondrial DNA structure were tracked through time by comparing ancient and modern populations of Eurasia. Analysis of haplogroup data included principal component analysis, multidimensional scaling, fixation index computation and genetic distance mapping. Haplotypic data were compared by haplotype sharing analysis, phylogenetic networks, Analysis of the Molecular Variance and coalescent simulations. The sequencing of a whole mitochondrial genome in a north east European Mesolithic individual lead to defining a new branch within the human mitochondrial tree. This work presents direct evidence that Mesolithic eastern Europeans belonged to the same Palaeolithic/Mesolithic genetic background as central and northern Europeans. It was also shown that prehistoric eastern Europeans were the recipients of multiple migrations from the East in prehistory that had not been previously detected and/or timed on the basis of modern mtDNA data. Ancient DNA also provided insights in the genetic history of European genetic outliers; the Saami, whose ancestral population still remain unidentified, and the Sardinians, whose genetic differentiation is proposed to be the result of mating isolation since at least the Bronze Age. This study demonstrates the power of aDNA to reveal previously unknown population processes in the genetic history of modern Eurasians.
Link



"feel free to comment on it if you discover any interesting nuggets in the text."
ReplyDeleteHere's a nugget for you: a couple of chapters are dealing with the discovery of mtDNA hg C1 at the 7,500 year old Mesolithic site of Uznyi Oleni Ostrov, Karelia (western Russia). Since C1 is predominantly an American Indian lineage and the back-migration of C1a to Siberia has already been proposed (http://www.plosone.org/article/info:doi/10.1371/journal.pone.0000829), this new finding provides further support for ancient Amerindian admixture in Europe (http://anthropogenesis.kinshipstudies.org/2012/09/how-europeans-got-to-be-10-american-indians/). The date of the find is old enough to exclude an Indo-European origin of European C1.
This find also dispels the idea that hg C1 detected in Icelanders represents recent (past 1000 years) Amerindian admixture. Icelandic and Karelian C1 likely belong to the same pool going back to the original peopling of Europe. Hence their low frequency in modern populations.
It also further supports my claim of Native American lineage through my mtDNA, the subclade of which is at least very, very closely related to X2a (the one "European" group documented to have be found among NA's). If Amerindian mtDNA of Mongoloid origin may be found in both ancient and modern Europe, then it follows that DNA of Caucasoid origin may also be found in ancient and modern North America, among certain lineages of some Native peoples (like, the Cherokee).
ReplyDeleteC1a is a sister clade, not a descendant of the American lineages C1b, C1c and C1d. Therefore we have no evidence about the back-migration from America, because C1 probably remained in Northeast Asia until the late birth of C1a (~8000 years; Derenko et al. 2010).
ReplyDeleteHey Dienekes what do you make of the presence of mt-DNA H in Mesolithic Karelians, namely individual UZOO-77 dating to 7500 ybp, with an HVR-I of 16311C, 16362C.
ReplyDeleteHere is Table-1 showing the clear presence of mt-DNA H in the site of Uznyi Oleni Ostrov:
http://i1133.photobucket.com/albums/m582/jeanlohizun/Sarkissianetal2011_Table-1.jpg
The Rostov Scythians appear like a mix of Eastern Europeans and Central Asians on those PCA plots. That's why they're so similar to Tatars.
ReplyDeleteThere's nothing particularly Southern Caucasoid about them. It's just that they're dragged towards Central Asia, which happens to be located to the lower right of the main European cluster.
Indeed, the distances between these Scythians and Georgians, Armenians, Kurds, Iranians and Pakistanis are very telling. There's no way they originated anywhere near the Caucasus or South Asia.
There's nothing particularly Southern Caucasoid about them. It's just that they're dragged towards Central Asia, which happens to be located to the lower right of the main European cluster.
ReplyDelete"Northern Caucasoids" are people like PWC or early Bronze Age Altai (the people at the top of the plot).
"Northern Europeans" were formed by admixture of northern Caucasoids with southern Caucasoids (see Skoglund et al.). Coon had the basic idea right when he postulated that: (i) Upper Paleolithic "survivals" were highest in northern Europe, but (ii) Europeans were formed by what he called "Mediterraneans" interacting with UP survivors.
The Scythians studied here do not appear to be northern Caucasoids. Of the unmixed Europeans they appear closest to Bulgarians and Albanians (p. 200) who are not even northern Europeans, let alone northern Caucasoids. And, they most certainly appear more "southern" than the earlier groups, consistent with other Iron Age groups.
Hey Dienekes what do you make of the presence of mt-DNA H in Mesolithic Karelians, namely individual UZOO-77 dating to 7500 ybp, with an HVR-I of 16311C, 16362C.
ReplyDeleteIt would have been nice if it were better resolved, so it's not clear what it's origin is. For the time being we can assume that some haplogroup H had reached NE Europe by 5.5ky BC, although the mechanism whereby this lineage reached levels of its present-day dominance are still obscure. I used to consider the possibility of natural selection, but the fact that H was already dominant in Copper Age groups like Bell Beakers suggests to me that a demographic reason might be more likely, although the precise routes whereby H entered Europe remain to be elucidated.
mt-DNA Haplogroup H appears to be dominant in early Neolithic sites from the Basque Country from which a significant sample is present. For example:
ReplyDelete-The early farming site of Los Cascajos (Navarre) dating back to 6185-5185 ybp shows 12/27 or 44.44% mt-DNA H, 8/27 or 29.63% mt-DNA U, 3/27 or 11.11% mt-DNA K1a , 2/27or 0.074% mt-DNA J, 1/27 or 3.7% mt-DNA T2, and 1/27 or 3.7% mt-DNA X.
-The early farming site of Paternanbidea (Navarre)dating back to 6090-5960 ybp shows 5/9 or 55.55% mt-DNA H, out of which 2/9 or 22.225 is mt-DNA H3, 1/9 or 1.11% mt-DNA U, 1/9 or 11.11% mt-DNA K , 1/9 or 11.11% mt-DNA I, and 1/9 or 11.11% mt-DNA HV.
The Mesolithic samples are far too small, but consist of the Magdalenian sample of Erralla, Guipuzcoa dating back to 12310 ybp and having mt-DNA U5, the sample of Linazeta cave, Deba, Guipuzcoa dated to 6230-6100 cal. BCE and having mt-DNA H(xH1, H3), and the site of Aizpea Navarra, dating back to 6600 ybp and having mt-DNA U5b1.
References:
http://www.plosone.org/article/info:doi/10.1371/journal.pone.0034417?imageURI=info:doi/10.1371/journal.pone.0034417.t001
http://thesesups.ups-tlse.fr/1392/1/2011TOU30177.pdf
Bronze Age Kazakhstan (KAZ-BA) appear to be southern Europeoid, and, also, noticeably more "West Eurasian" than the others
ReplyDeleteIt is interesting that apparently there was a southern European-like population in the Central Asian heartland during the Bronze Age. It seems to have been largely replaced by West Asian, East Eurasian and South Asian leaning populations during the early Iron Age.
Jaska - C1a is a descendant of C1. There are no known C1b, C1c or C1d sequences in Asia and so no evidence that the mutations defining these branches occurred there.
ReplyDeleteThe estimated age of C1a, according to Behar et al. 2012, was only 4837 years and the entry point could easily have been Japan based upon 1) proximity and 2) because Japanese 'C' sequences, pretty much, only belong to C1a (6/828 - C1a, 1/828 - C4a1)
@Jaska
ReplyDelete"Therefore we have no evidence about the back-migration from America, because C1 probably remained in Northeast Asia until the late birth of C1a (~8000 years; Derenko et al. 2010)."
But Amerindian C1b,c,d are much more diverse, more frequent and date older than Siberian C1a. European C1e is also no match to Amerindian C1b,c, d. By all standards accepted in genetic literature (compare, for example, mtDNA M1, which is considered to have back-migrated from Asia, is less frequent, less diverse and dates younger than Asian M lineages, although technically they are sister clades.
A back migration from the New World is better supported by interdisciplinary evidence than the founding migration into the New World. See more at http://anthropogenesis.kinshipstudies.org/2012/11/yuzhnyi-olenii-ostrov-ancient-mtdna-evidence-for-amerindian-admixture-in-europe/
Looking at the Pre-Iron-Age PC plot, the Rostov Scythians look about midway between the Middle East (Saudis, Iraqis, etc.) and the Ancient Eurasians (Bronze Age Altai, Pitted Ware, etc).
ReplyDeleteAlmost as if they were a mix of those 2 types (Middle Eastern and Proto-Europoid).
PWC and ALT-BA look _way_ northerly. There is as much distance between them and FIN as between FIN and GRC. Are these ancient nordic CM? The modern norse may look like Meds in comparison. It could be spectacular to see what they looked like. It would be interesting to see how far northward individual modern norse might tend.
ReplyDeleteIt is interesting that the S_E_ European populations tend westward of northern Euro and PWC. It implies that the eastward shift in the north is ancient and that modern populations have been pulled westward by mixing with southern EUropean/ west Asian groups that lack eastward shift. One could draw a diagonal line from Cyprus to PWC/ ALT-BA with modern Europe, from south to north, falling along on the line.
(I admit that I havent read the 200 page thesis and that I am just looking at the pictures lol. I have no idea how they did a PCA from mtDNA and how it might accord with autosomes)
Jean Manco did not post this data on her site, she must have quit
ReplyDelete