This is an open access paper describing the phase1 data of the 1000 Genomes Project. There is plenty of interest in the paper and supplement, but look at Figure S8 (left). This indicates the median shared haplotype length around f2 sites, i.e., sites where the variant exists twice in the sample and hence it makes sense to speak of shared length.
The maximum such length is for FIN (Finns) at 140kb, but it seems fairly obvious visually that the lowest sharing is found in multi-origin populations from the Americas (MXL, CLM, PUR, AS), in which segments are probably "interrupted" because of admixture. African populations (LWK/YRI) also tend to have low sharing, followed by Europeans, and East Asians.
There are little details in evidence: for example, IBS sharing with Luhya (LWK) seems higher than the European average, consistent with some level of African admixture in Spain, that has probably contributed some African haplotypes.
There seems to be a hint of an excess of sharing between Japanese (JPT) and Luhya (LWK). I have to wonder whether this might have something to do with Y-haplogroup D which links the Japanese with African Y-haplogroup E bearers. An excess of sharing between CHS (Singapore Chinese) and PUR (Puerto Rico) also seems to be suggested, for which I have no good hypothesis.
Nature 491, 56–65 (01 November 2012) doi:10.1038/nature11632
An integrated map of genetic variation from 1,092 human genomes
The 1000 Genomes Project Consortium
By characterizing the geographic and functional spectrum of human genetic variation, the 1000 Genomes Project aims to build a resource to help to understand the genetic contribution to disease. Here we describe the genomes of 1,092 individuals from 14 populations, constructed using a combination of low-coverage whole-genome and exome sequencing. By developing methods to integrate information across several algorithms and diverse data sources, we provide a validated haplotype map of 38|[thinsp]|million single nucleotide polymorphisms, 1.4|[thinsp]|million short insertions and deletions, and more than 14,000 larger deletions. We show that individuals from different populations carry different profiles of rare and common variants, and that low-frequency variants show substantial geographic differentiation, which is further increased by the action of purifying selection. We show that evolutionary conservation and coding consequence are key determinants of the strength of purifying selection, that rare-variant load varies substantially across biological pathways, and that each individual contains hundreds of rare non-coding variants at conserved sites, such as motif-disrupting changes in transcription-factor-binding sites. This resource, which captures up to 98% of accessible single nucleotide polymorphisms at a frequency of 1% in related populations, enables analysis of common and low-frequency variants in individuals from diverse, including admixed, populations.
Link
October 31, 2012
mtDNA of Bronze Age pastoral nomads of Ukraine
This is just a presentation, so it will be interesting to find an alternative source for it. Still, it seems to agree with other evidence about the hybrid origin of Bronze Age European steppe nomads. A detailed look at the evidence from the Balkans, north Pontic steppe, and the Caucasus (and perhaps also the trans-Caspian region) will determine on who went where and when.
Genetic Analysis of Ancient Human Remains from the Bronze Age Nomadic Steppe Cultures of Ukraine
Jeff Pashnick, Grand Valley State University
During the transition between the late Neolithic and the Early Bronze Age (EBA) proto-Indo-European languages began to spread from southeastern steppes (prairielands) westwards into Europe. Southern Ukraine (North Pontic Region, NPR) was the meeting place between the Old Europe and steppe nomadic cultures. Using mitochondrial DNA (mtDNA), we tested ancient human remains from the EBA cultures from the NPR to determine if there was genetic evidence for the mingling of these cultures. Our data shows mtDNA lineages (haplogroups) of nomadic pastoralists in the NPR to have mainly common haplogroups with European hunter-gatherer cultures, with an inclusion of haplogroups common to farming cultures of Europe. The similarities in the haplogroup composition between European Neolithic hunter-gatherers and the NPR steppe pastoralists suggests that they share a common genetic past, in part influenced by the neighboring farmers and in part stemming from the Mesolithic native European ancestry.
Link
Genetic Analysis of Ancient Human Remains from the Bronze Age Nomadic Steppe Cultures of Ukraine
Jeff Pashnick, Grand Valley State University
During the transition between the late Neolithic and the Early Bronze Age (EBA) proto-Indo-European languages began to spread from southeastern steppes (prairielands) westwards into Europe. Southern Ukraine (North Pontic Region, NPR) was the meeting place between the Old Europe and steppe nomadic cultures. Using mitochondrial DNA (mtDNA), we tested ancient human remains from the EBA cultures from the NPR to determine if there was genetic evidence for the mingling of these cultures. Our data shows mtDNA lineages (haplogroups) of nomadic pastoralists in the NPR to have mainly common haplogroups with European hunter-gatherer cultures, with an inclusion of haplogroups common to farming cultures of Europe. The similarities in the haplogroup composition between European Neolithic hunter-gatherers and the NPR steppe pastoralists suggests that they share a common genetic past, in part influenced by the neighboring farmers and in part stemming from the Mesolithic native European ancestry.
Link
Improved phylogenetic resolution within Y-haplogroup R1a1
Here is the table of haplogroup frequencies:
I have written before that I envision R1a1 to have been anciently distributed in the wide arc of "flatlands" north and east of the Caspian sea, complementing R-M269 whose distribution is suggestive of the short arc of "highlands" west and south of it.
The current distribution is strongly geographically bimodal, with peaks in eastern Europe and South Asia. The phylogeny of this group is continuously refined, but one of the problems with the "commercial" studies of haplogroups is that they tend to consist of samples drawn primarily of the groups of people who are likely to have heard of DNA testing, and this excludes large regions within the Eurasian heartland. For example, Z280 is listed as "Central and Eastern Europe Western Asia" in the R1a1a and subclades project, but here makes up 2/9 R-M198 related samples from the Central Asian Uzbek sample. Similarly, M458 is listed as Central Europe, but occurs in 1/9 Uzbek. We can't know for sure which SNP occurs where until we test large representative samples.
There are various aspects of the problem that need to be considered: the absence of R1a-related lineages in pre-Copper Age Europe, and of the distant R1b relative, together with the firm rooting of the R1 clade in Asia indicate that the lineage leading to R1a1 traces its ancestry to a migration into Europe. How and when that migration occurred is an open problem. The paucity of SNP diversity in South Asia, at least in the available samples, indicates that a migration into South Asia also occurred. So, I agree with the authors "that an early differentiation zone of R1a1-M198 conceivably occurred somewhere within the Eurasian Steppes or the Middle East and Caucasus region as they lie between South Asia and Eastern Europe."
My working hypothesis is that the bimodal distribution of R1a1-related lineages in Eurasia can be explained on the basis of two expansions involving largely Z283 in Europe and Z93 in Asia. The source of those expansions may have been Central Asia, and the relative scarcity of R1a1 in that region (relative to Europe and South Asia) may be the result of a subsequent movement of East Eurasians into it, at the same time as the expansion of Altaic speakers. The Uzbek sample in this paper give us a strong hint about the existence of an overlap zone in Central Asia, but the SNP diversity is little studied in the populations of the -stan states, and ancient DNA samples are missing.
The issue of time depth is also relevant, as it will anchor in time the evolutionary relationships between different populations of R1a1 descendants. This can be achieved both by (i) typing ancient samples for the relevant markers, which will provide -assuming a positive result- a terminus ante quem for the appearance of particular SNP, and (ii) sequencing modern Y-chromosomes to determine their TMRCA.
AJPA DOI: 10.1002/ajpa.22167
Brief communication: New Y-chromosome binary markers improve phylogenetic resolution within haplogroup R1a1
Horolma Pamjav et al.
Abstract
Haplogroup R1a1-M198 is a major clade of Y chromosomal haplogroups which is distributed all across Eurasia. To this date, many efforts have been made to identify large SNP-based subgroups and migration patterns of this haplogroup. The origin and spread of R1a1 chromosomes in Eurasia has, however, remained unknown due to the lack of downstream SNPs within the R1a1 haplogroup. Since the discovery of R1a1-M458, this is the first scientific attempt to divide haplogroup R1a1-M198 into multiple SNP-based sub-haplogroups. We have genotyped 217 R1a1-M198 samples from seven different population groups at M458, as well as the Z280 and Z93 SNPs recently identified from the “1000 Genomes Project”.
The two additional binary markers present an effective tool because now more than 98% of the samples analyzed assign to one of the three sub-haplogroups. R1a1-M458 and R1a1-Z280 were typical for the Hungarian population groups, whereas R1a1-Z93 was typical for Malaysian Indians and the Hungarian Roma. Inner and Central Asia is an overlap zone for the R1a1-Z280 and R1a1-Z93 lineages. This pattern implies that an early differentiation zone of R1a1-M198 conceivably occurred somewhere within the Eurasian Steppes or the Middle East and Caucasus region as they lie between South Asia and Eastern Europe. The detection of the Z93 paternal genetic imprint in the Hungarian Roma gene pool is consistent with South Asian ancestry and amends the view that H1a-M82 is their only discernible paternal lineage of Indian heritage.
Link
I have written before that I envision R1a1 to have been anciently distributed in the wide arc of "flatlands" north and east of the Caspian sea, complementing R-M269 whose distribution is suggestive of the short arc of "highlands" west and south of it.
The current distribution is strongly geographically bimodal, with peaks in eastern Europe and South Asia. The phylogeny of this group is continuously refined, but one of the problems with the "commercial" studies of haplogroups is that they tend to consist of samples drawn primarily of the groups of people who are likely to have heard of DNA testing, and this excludes large regions within the Eurasian heartland. For example, Z280 is listed as "Central and Eastern Europe Western Asia" in the R1a1a and subclades project, but here makes up 2/9 R-M198 related samples from the Central Asian Uzbek sample. Similarly, M458 is listed as Central Europe, but occurs in 1/9 Uzbek. We can't know for sure which SNP occurs where until we test large representative samples.
There are various aspects of the problem that need to be considered: the absence of R1a-related lineages in pre-Copper Age Europe, and of the distant R1b relative, together with the firm rooting of the R1 clade in Asia indicate that the lineage leading to R1a1 traces its ancestry to a migration into Europe. How and when that migration occurred is an open problem. The paucity of SNP diversity in South Asia, at least in the available samples, indicates that a migration into South Asia also occurred. So, I agree with the authors "that an early differentiation zone of R1a1-M198 conceivably occurred somewhere within the Eurasian Steppes or the Middle East and Caucasus region as they lie between South Asia and Eastern Europe."
My working hypothesis is that the bimodal distribution of R1a1-related lineages in Eurasia can be explained on the basis of two expansions involving largely Z283 in Europe and Z93 in Asia. The source of those expansions may have been Central Asia, and the relative scarcity of R1a1 in that region (relative to Europe and South Asia) may be the result of a subsequent movement of East Eurasians into it, at the same time as the expansion of Altaic speakers. The Uzbek sample in this paper give us a strong hint about the existence of an overlap zone in Central Asia, but the SNP diversity is little studied in the populations of the -stan states, and ancient DNA samples are missing.
The issue of time depth is also relevant, as it will anchor in time the evolutionary relationships between different populations of R1a1 descendants. This can be achieved both by (i) typing ancient samples for the relevant markers, which will provide -assuming a positive result- a terminus ante quem for the appearance of particular SNP, and (ii) sequencing modern Y-chromosomes to determine their TMRCA.
AJPA DOI: 10.1002/ajpa.22167
Brief communication: New Y-chromosome binary markers improve phylogenetic resolution within haplogroup R1a1
Horolma Pamjav et al.
Abstract
Haplogroup R1a1-M198 is a major clade of Y chromosomal haplogroups which is distributed all across Eurasia. To this date, many efforts have been made to identify large SNP-based subgroups and migration patterns of this haplogroup. The origin and spread of R1a1 chromosomes in Eurasia has, however, remained unknown due to the lack of downstream SNPs within the R1a1 haplogroup. Since the discovery of R1a1-M458, this is the first scientific attempt to divide haplogroup R1a1-M198 into multiple SNP-based sub-haplogroups. We have genotyped 217 R1a1-M198 samples from seven different population groups at M458, as well as the Z280 and Z93 SNPs recently identified from the “1000 Genomes Project”.
The two additional binary markers present an effective tool because now more than 98% of the samples analyzed assign to one of the three sub-haplogroups. R1a1-M458 and R1a1-Z280 were typical for the Hungarian population groups, whereas R1a1-Z93 was typical for Malaysian Indians and the Hungarian Roma. Inner and Central Asia is an overlap zone for the R1a1-Z280 and R1a1-Z93 lineages. This pattern implies that an early differentiation zone of R1a1-M198 conceivably occurred somewhere within the Eurasian Steppes or the Middle East and Caucasus region as they lie between South Asia and Eastern Europe. The detection of the Z93 paternal genetic imprint in the Hungarian Roma gene pool is consistent with South Asian ancestry and amends the view that H1a-M82 is their only discernible paternal lineage of Indian heritage.
Link
October 29, 2012
Autozygosity and the 1.2x10-8 mut/bp/gen mutation rate
A number of different methods have converged on the 1.2x10-8 mutation rate, or slightly slower/faster rates around this value. In the current paper, the authors exploited inbreeding within the Hutterite population to identify segments of autozygous DNA, i.e., chunks of DNA where the two copies in an individual were inherited from the same ancestor, but via different genealogical paths. These copies are nearly-identical, but not entirely so, since they followed different sequences of meioses in different bodies on their way from the common ancestor to the modern individuals. By counting these differences and dividing by the number of intervening meioses, an estimate of the mutation rate can be arrived at.
Nature Genetics 44, 1277–1281 (2012) doi:10.1038/ng.2418
Estimating the human mutation rate using autozygosity in a founder population
Catarina D Campbell et al.
Knowledge of the rate and pattern of new mutation is critical to the understanding of human disease and evolution. We used extensive autozygosity in a genealogically well-defined population of Hutterites to estimate the human sequence mutation rate over multiple generations. We sequenced whole genomes from 5 parent-offspring trios and identified 44 segments of autozygosity. Using the number of meioses separating each pair of autozygous alleles and the 72 validated heterozygous single-nucleotide variants (SNVs) from 512 Mb of autozygous DNA, we obtained an SNV mutation rate of 1.20 × 10−8 (95% confidence interval 0.89–1.43 × 10−8) mutations per base pair per generation. The mutation rate for bases within CpG dinucleotides (9.72 × 10−8) was 9.5-fold that of non-CpG bases, and there was strong evidence (P = 2.67 × 10−4) for a paternal bias in the origin of new mutations (85% paternal). We observed a non-uniform distribution of heterozygous SNVs (both newly identified and known) in the autozygous segments (P = 0.001), which is suggestive of mutational hotspots or sites of long-range gene conversion.
Link
Nature Genetics 44, 1277–1281 (2012) doi:10.1038/ng.2418
Estimating the human mutation rate using autozygosity in a founder population
Catarina D Campbell et al.
Knowledge of the rate and pattern of new mutation is critical to the understanding of human disease and evolution. We used extensive autozygosity in a genealogically well-defined population of Hutterites to estimate the human sequence mutation rate over multiple generations. We sequenced whole genomes from 5 parent-offspring trios and identified 44 segments of autozygosity. Using the number of meioses separating each pair of autozygous alleles and the 72 validated heterozygous single-nucleotide variants (SNVs) from 512 Mb of autozygous DNA, we obtained an SNV mutation rate of 1.20 × 10−8 (95% confidence interval 0.89–1.43 × 10−8) mutations per base pair per generation. The mutation rate for bases within CpG dinucleotides (9.72 × 10−8) was 9.5-fold that of non-CpG bases, and there was strong evidence (P = 2.67 × 10−4) for a paternal bias in the origin of new mutations (85% paternal). We observed a non-uniform distribution of heterozygous SNVs (both newly identified and known) in the autozygous segments (P = 0.001), which is suggestive of mutational hotspots or sites of long-range gene conversion.
Link
Assessment of ancient European DNA with 'globe13'
Here is my assessment of ancient DNA from Europe using the globe13 calculator:
You can consult the spreadsheet for the distribution of these components in modern populations. As in previous analyses, the main distinction is between Northern European-like Mesolithic population (Ajv52, Ajv70, and Bra1), and Mediterranean-like Neolithic (Oetzi and Gok4) one.
You can consult the spreadsheet for the distribution of these components in modern populations. As in previous analyses, the main distinction is between Northern European-like Mesolithic population (Ajv52, Ajv70, and Bra1), and Mediterranean-like Neolithic (Oetzi and Gok4) one.
October 28, 2012
October 27, 2012
Inter-relationships between 'world' components
In a previous post I calculated f3-statistics between my K=7 and K=12 ancestral components. The basic idea is to discover which component A can be seen as a mixture of two other components, B and C, in which case (assuming A does not have excessive drift), we expect a negative f3(A; B, C) statistic.
As part of my analysis of the world dataset, I calculated f3-statistics for each of the K=3 to K=12, that is, for some K, I tried to see if one of the K inferred components could be seen as a mixture of the remaining K-1. It turns out that no negative f3 statistics appeared at all, and this suggests that the components inferred by ADMIXTURE at each K tend to form an "orthogonal" set that are not mixtures of each other.
More generally, we can calculate f3 statistics where A, B, and C are components inferred from any of the K=3 to K=12 runs. There is a total of 75 such components, and hence 75*(74 choose 2) = 202,575 such f3 statistics. Since calculating these would take a while (and would become intractable as K increases further), I decided to calculate pairwise f3 statistics, i.e., statistics where A, B, and C are constrained to be from successive K, K+1 runs. The significant results can be seen in the spreadsheet.
It might be worthwhile to develop an automated way of using these statistics to guide us in the interpretation of ADMIXTURE components. But, they are useful, in any case, as a source of information.
For example, consider the following (the third column represents the mixed population):
Atlantic_Baltic_6/globe6_Z Near_East_6/globe6_Z European_5/globe5_Z -0.013911 0.000084 -166.457
This means that the European component at K=5 can be seen as a mix of the Atlantic_Baltic and Near_East components at K=6. So, this suggests that the European component can be seen as "secondary", the product of admixture. But:
European_5/globe5_Z Amerindian_5/globe5_Z Atlantic_Baltic_6/globe6_Z -0.003964 0.000175 -22.588
This indicates conversely that the Atlantic_Baltic at K=6 component can be seen as a mix of the European and Amerindian components at K=6.
It would be very interesting to use f-statistics to guide one in the choice of an "orthogonal" set of ancestral populations, or to summarize the relationships between them in tree or network form. One could potentially use my ADMIXTURE to TreeMix script to do something like this, although as K increases, there is a combinatorial explosion in the total number of components with a probable runtime slowdown/memory usage blowup which might render this approach unusable, at least for large K.
As part of my analysis of the world dataset, I calculated f3-statistics for each of the K=3 to K=12, that is, for some K, I tried to see if one of the K inferred components could be seen as a mixture of the remaining K-1. It turns out that no negative f3 statistics appeared at all, and this suggests that the components inferred by ADMIXTURE at each K tend to form an "orthogonal" set that are not mixtures of each other.
More generally, we can calculate f3 statistics where A, B, and C are components inferred from any of the K=3 to K=12 runs. There is a total of 75 such components, and hence 75*(74 choose 2) = 202,575 such f3 statistics. Since calculating these would take a while (and would become intractable as K increases further), I decided to calculate pairwise f3 statistics, i.e., statistics where A, B, and C are constrained to be from successive K, K+1 runs. The significant results can be seen in the spreadsheet.
It might be worthwhile to develop an automated way of using these statistics to guide us in the interpretation of ADMIXTURE components. But, they are useful, in any case, as a source of information.
For example, consider the following (the third column represents the mixed population):
Atlantic_Baltic_6/globe6_Z Near_East_6/globe6_Z European_5/globe5_Z -0.013911 0.000084 -166.457
This means that the European component at K=5 can be seen as a mix of the Atlantic_Baltic and Near_East components at K=6. So, this suggests that the European component can be seen as "secondary", the product of admixture. But:
European_5/globe5_Z Amerindian_5/globe5_Z Atlantic_Baltic_6/globe6_Z -0.003964 0.000175 -22.588
This indicates conversely that the Atlantic_Baltic at K=6 component can be seen as a mix of the European and Amerindian components at K=6.
It would be very interesting to use f-statistics to guide one in the choice of an "orthogonal" set of ancestral populations, or to summarize the relationships between them in tree or network form. One could potentially use my ADMIXTURE to TreeMix script to do something like this, although as K increases, there is a combinatorial explosion in the total number of components with a probable runtime slowdown/memory usage blowup which might render this approach unusable, at least for large K.
October 26, 2012
IBD length distribution and demographic history (Palamara)
Another interesting new paper in AJHG deals with the problem of inferring the demographic history of a population from the length distribution of IBD segments. I wonder how admixture (which is likely to have occurred in both chosen real-world examples used in the paper, the Ashkenazi Jews and the Maasai) may affect the accuracy of the reconstructed demographic history.
In any case, the conclusions are worth mentioning in themselves. For the Ashkenazi:
The American Journal of Human Genetics, 25 October 2012 doi:10.1016/j.ajhg.2012.08.030
Length Distributions of Identity by Descent Reveal Fine-Scale Demographic History
Pier Francesco Palamara et al.
Data-driven studies of identity by descent (IBD) were recently enabled by high-resolution genomic data from large cohorts and scalable algorithms for IBD detection. Yet, haplotype sharing currently represents an underutilized source of information for population-genetics research. We present analytical results on the relationship between haplotype sharing across purportedly unrelated individuals and a population’s demographic history. We express the distribution of IBD sharing across pairs of individuals for segments of arbitrary length as a function of the population’s demography, and we derive an inference procedure to reconstruct such demographic history. The accuracy of the proposed reconstruction methodology was extensively tested on simulated data. We applied this methodology to two densely typed data sets: 500 Ashkenazi Jewish (AJ) individuals and 56 Kenyan Maasai (MKK) individuals (HapMap 3 data set). Reconstructing the demographic history of the AJ cohort, we recovered two subsequent population expansions, separated by a severe founder event, consistent with previous analysis of lower-throughput genetic data and historical accounts of AJ history. In the MKK cohort, high levels of cryptic relatedness were detected. The spectrum of IBD sharing is consistent with a demographic model in which several small-sized demes intermix through high migration rates and result in enrichment of shared long-range haplotypes. This scenario of historically structured demographies might explain the unexpected abundance of runs of homozygosity within several populations.
Link
In any case, the conclusions are worth mentioning in themselves. For the Ashkenazi:
We obtained an improved fit for a population composed of ~2,300 ancestors 200 generations before the present; this population exponentially expanded to reach ~45,000 individuals 34 generations ago. After a severe founder event, the population was reduced to ~270 individuals, which then expanded rapidly during 33 generations (rate r ~ 0.29) and reached a modern population of ~4,300,000 individuals.And, for the Maasai:
Optimizing a model of exponential expansion and contraction (Figure 1A), we obtained a good fit to the observed IBD frequency spectrum (Figure 6), suggesting that an ancestral population of ~23,500 individuals decreased to ~500 current individuals during the course of 23 generations (r ~ -0.17). We note that this result might not be driven by an actual gradual population contraction in the MKK individuals, but it most likely reflects the societal structure of this seminomadic population. ... We thus used the village model to analyze the MKK demography and relied on coalescent simulations to retrieve its parameters: migration rate, size, and number of villages that provide a good fit for the empirical distribution of IBD segments.We observed a compatible fit for this model, in which 44 villages of 485 individuals each intermix with a migration rate of 0.13 individuals per generation (Figure 6).If I understand this correctly, it appears that Maasai (MKK) individuals share long IBD segments not because their population has contracted (and hence they're all descended from a limited number of founders, as is the case for Ashkenazi Jews), but rather because their social structure follows the "village model" in which people share shallow ancestry (and hence long IBD) with other people in their "village" and exchange genes with other "villages".
The American Journal of Human Genetics, 25 October 2012 doi:10.1016/j.ajhg.2012.08.030
Length Distributions of Identity by Descent Reveal Fine-Scale Demographic History
Pier Francesco Palamara et al.
Data-driven studies of identity by descent (IBD) were recently enabled by high-resolution genomic data from large cohorts and scalable algorithms for IBD detection. Yet, haplotype sharing currently represents an underutilized source of information for population-genetics research. We present analytical results on the relationship between haplotype sharing across purportedly unrelated individuals and a population’s demographic history. We express the distribution of IBD sharing across pairs of individuals for segments of arbitrary length as a function of the population’s demography, and we derive an inference procedure to reconstruct such demographic history. The accuracy of the proposed reconstruction methodology was extensively tested on simulated data. We applied this methodology to two densely typed data sets: 500 Ashkenazi Jewish (AJ) individuals and 56 Kenyan Maasai (MKK) individuals (HapMap 3 data set). Reconstructing the demographic history of the AJ cohort, we recovered two subsequent population expansions, separated by a severe founder event, consistent with previous analysis of lower-throughput genetic data and historical accounts of AJ history. In the MKK cohort, high levels of cryptic relatedness were detected. The spectrum of IBD sharing is consistent with a demographic model in which several small-sized demes intermix through high migration rates and result in enrichment of shared long-range haplotypes. This scenario of historically structured demographies might explain the unexpected abundance of runs of homozygosity within several populations.
Link
October 25, 2012
Instantaneous vs. continuous admixture dynamics (Jin et al. 2012)
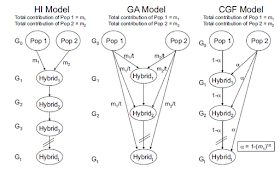
A new paper in AJHG discusses the distribution of chromosomal segments of distinct ancestry (CSDAs) under three different models of admixture dynamics (left). In the hybrid isolation (HI) model, admixture is instantaneous and results in a hybrid population that evolves with drift and recombination only. In the gradual admixture (GA) model, the hybrid population continues to receive admixture from the unadmixed parental populations. Finally, in the continuous gene flow model (CGF), one of the populations becomes admixed while the other continues to exist unadmixed and to contribute to the admixed one.
In practical terms, the HI model results in the diminution of CSDA length due to recombination over time, and at "present" there is a paucity of long CSDAs. In the GA model there are more long CSDAs for both populations, while in the CGF model there is an asymmetry in the CSDAs donated by Pop1 and Pop2, with those from the "donor" population being longer (because fresh "long" segments are added in every generation).
The conclusions of the paper regarding some particular admixture cases are also interesting. For African Americans:
I should mention that according to HAPMIX, the admixture time was 7 generations, with is close to the 6 +/- 1 generations inferred by rolloff analysis by Moorjani et al. So, in this case this admixture time appears to be an "average" of a continuing process of admixture that began 14 generations ago.
Onto Mexicans:
On the African admixture in Mozabites:
A similar pattern was evident for Bedouin, Palestinians, and Druze:
The American Journal of Human Genetics, 25 October 2012 doi:10.1016/j.ajhg.2012.09.008
Exploring Population Admixture Dynamics via Empirical and Simulated Genome-Wide Distribution of Ancestral Chromosomal Segments
Wenfei Jin et al
Abstract
The processes of genetic admixture determine the haplotype structure and linkage disequilibrium patterns of the admixed population, which is important for medical and evolutionary studies. However, most previous studies do not consider the inherent complexity of admixture processes. Here we proposed two approaches to explore population admixture dynamics, and we demonstrated, by analyzing genome-wide empirical and simulated data, that the approach based on the distribution of chromosomal segments of distinct ancestry (CSDAs) was more powerful than that based on the distribution of individual ancestry proportions. Analysis of 1,890 African Americans showed that a continuous gene flow model, in which the African American population continuously received gene flow from European populations over about 14 generations, best explained the admixture dynamics of African Americans among several putative models. Interestingly, we observed that some African Americans had much more European ancestry than the simulated samples, indicating substructures of local ancestries in African Americans that could have been caused by individuals from some particular lineages having repeatedly admixed with people of European ancestry. In contrast, the admixture dynamics of Mexicans could be explained by a gradual admixture model in which the Mexican population continuously received gene flow from both European and Amerindian populations over about 24 generations. Our results also indicated that recent gene flows from Sub-Saharan Africans have contributed to the gene pool of Middle Eastern populations such as Mozabite, Bedouin, and Palestinian. In summary, this study not only provides approaches to explore population admixture dynamics, but also advances our understanding on population history of African Americans, Mexicans, and Middle Eastern populations.
Link
In practical terms, the HI model results in the diminution of CSDA length due to recombination over time, and at "present" there is a paucity of long CSDAs. In the GA model there are more long CSDAs for both populations, while in the CGF model there is an asymmetry in the CSDAs donated by Pop1 and Pop2, with those from the "donor" population being longer (because fresh "long" segments are added in every generation).
The conclusions of the paper regarding some particular admixture cases are also interesting. For African Americans:
Although the actual population admixture of African Americans might be more complex than what our simulation suggested, the CGF1 model setting at 14 generations was found to be reasonably epresentative, capturing the main pattern of the population admixture dynamics.The CGF1 model has Africans as recipients and Europeans as donors. This makes sense, since African Americans are descended from slaves who were transported to the New World, with the slave trade ending centuries ago, hence there was mostly no replenishment of the AA population with fresh African-origin individuals. On the other hand, European Americans, both due to social dynamics and their numerical majority continued to exist as a distinct population that contributed to the AA population.
I should mention that according to HAPMIX, the admixture time was 7 generations, with is close to the 6 +/- 1 generations inferred by rolloff analysis by Moorjani et al. So, in this case this admixture time appears to be an "average" of a continuing process of admixture that began 14 generations ago.
Onto Mexicans:
In short, the GA model at 24 generations fit the empirical data best among all these simulated scenarios, as indicated by the distribution of EMDs.Again, this makes sense, because in Mexico there continued to exist unadmixed populations of Europeans and Amerindians that contributed to the Mestizo population of the country.
On the African admixture in Mozabites:
Comparing the empirical distribution of CSDAs with that simulated, we found that the Mozabite admixture process essentially fit the HI model with 100 generations since admixture. There was an almost complete absence of recent gene flow from European populations to the Mozabite gene pool (Figure 6A). For the Sub-Saharan African ancestral component, there were more long CSDAs at the tail of empirical distribution than those in the HI model, which confirmed that recent gene flow from African populations had contributed to the Mozabite gene pool (Figure 6B).Again, this makes sense: Berber groups were not replenished from other Caucasoid sources, so their original admixture with native Africans resulted in a blend that persisted largely unaffected by "Europeans", but did find occasion of admixture with Sub-Saharans. Hence, the asymmetry in the presence of long "European" vs. "Sub-Saharan" segments.
A similar pattern was evident for Bedouin, Palestinians, and Druze:
Analyses of European ancestral component in Bedouin and Palestinian populations also showed that the empirical distributions essentially fit the HI model for both populations (Figures 6C and 6E). Although the empirical CSDA distribution of Sub-Saharan African ancestral component also fit the HI model best, both distributions showed a long tail at the right compared with those under the HI model, indicating that recent gene flow from Sub-Saharan Africans also contributed to the two admixed populations (Figures 6D and 6F). ... For Druze, their European component of ancestry fit the HI model very well. However, their African ancestral component contained much shorter CSDAs than those of simulated (Figure S14), which might indicate that previous studies had underestimated the admixture time of Druze. In addition, populations receiving recent gene flow from their parental populations showed higher variation of individual ancestral proportions than those who did not (Figure S13).The Druze have well-known Egyptian connections, and they may have largely avoided Sub-Saharan African admixture during the Islamic period, principally because of its avoidance of proselytism. Hence, their African admixture may stem from Egyptian adherents who were themselves a product of much earlier Caucasoid/Sub-Saharan admixture during the course of pre-Islamic Egypt.
The American Journal of Human Genetics, 25 October 2012 doi:10.1016/j.ajhg.2012.09.008
Exploring Population Admixture Dynamics via Empirical and Simulated Genome-Wide Distribution of Ancestral Chromosomal Segments
Wenfei Jin et al
Abstract
The processes of genetic admixture determine the haplotype structure and linkage disequilibrium patterns of the admixed population, which is important for medical and evolutionary studies. However, most previous studies do not consider the inherent complexity of admixture processes. Here we proposed two approaches to explore population admixture dynamics, and we demonstrated, by analyzing genome-wide empirical and simulated data, that the approach based on the distribution of chromosomal segments of distinct ancestry (CSDAs) was more powerful than that based on the distribution of individual ancestry proportions. Analysis of 1,890 African Americans showed that a continuous gene flow model, in which the African American population continuously received gene flow from European populations over about 14 generations, best explained the admixture dynamics of African Americans among several putative models. Interestingly, we observed that some African Americans had much more European ancestry than the simulated samples, indicating substructures of local ancestries in African Americans that could have been caused by individuals from some particular lineages having repeatedly admixed with people of European ancestry. In contrast, the admixture dynamics of Mexicans could be explained by a gradual admixture model in which the Mexican population continuously received gene flow from both European and Amerindian populations over about 24 generations. Our results also indicated that recent gene flows from Sub-Saharan Africans have contributed to the gene pool of Middle Eastern populations such as Mozabite, Bedouin, and Palestinian. In summary, this study not only provides approaches to explore population admixture dynamics, but also advances our understanding on population history of African Americans, Mexicans, and Middle Eastern populations.
Link
Wide-bodied early Holocene north Americans
Am J Phys Anthropol DOI: 10.1002/ajpa.22154
Skeletal variation among early holocene north american humans: Implications for origins and diversity in the americas
Benjamin M. Auerbach
The movement of humans into the Americas remains a major topic of debate among scientific disciplines. Central to this discussion is ascertaining the timing and migratory routes of the earliest colonizers, in addition to understanding their ancestry. Molecular studies have recently argued that the colonizing population was isolated from other Asian populations for an extended period before proceeding to colonize the Americas. This research has suggested that Beringia was the location of this “incubation,” though archaeological and skeletal data have not yet supported this hypothesis. This study employs the remains of the five most complete North American male early Holocene skeletons to examine patterns of human morphology at the earliest observable time period. Stature, body mass, body breadth, and limb proportions are examined in the context of male skeletal samples representing the range of morphological variation in North America in the last two millennia of the Holocene. These are also compared with a global sample. Results indicate that early Holocene males have variable postcranial morphologies, but all share the common trait of wide bodies. This trait, which is retained in more recent indigenous North American groups, is associated with adaptations to cold climates. Peoples from the Americas exhibit wider bodies than other populations sampled globally. This pattern suggests the common ancestral population of all of these indigenous American groups had reduced morphological variation in this trait. Furthermore, this provides support for a single, possibly high latitude location for the genetic isolation of ancestors of the human colonizers of the Americas.
Link
Skeletal variation among early holocene north american humans: Implications for origins and diversity in the americas
Benjamin M. Auerbach
The movement of humans into the Americas remains a major topic of debate among scientific disciplines. Central to this discussion is ascertaining the timing and migratory routes of the earliest colonizers, in addition to understanding their ancestry. Molecular studies have recently argued that the colonizing population was isolated from other Asian populations for an extended period before proceeding to colonize the Americas. This research has suggested that Beringia was the location of this “incubation,” though archaeological and skeletal data have not yet supported this hypothesis. This study employs the remains of the five most complete North American male early Holocene skeletons to examine patterns of human morphology at the earliest observable time period. Stature, body mass, body breadth, and limb proportions are examined in the context of male skeletal samples representing the range of morphological variation in North America in the last two millennia of the Holocene. These are also compared with a global sample. Results indicate that early Holocene males have variable postcranial morphologies, but all share the common trait of wide bodies. This trait, which is retained in more recent indigenous North American groups, is associated with adaptations to cold climates. Peoples from the Americas exhibit wider bodies than other populations sampled globally. This pattern suggests the common ancestral population of all of these indigenous American groups had reduced morphological variation in this trait. Furthermore, this provides support for a single, possibly high latitude location for the genetic isolation of ancestors of the human colonizers of the Americas.
Link
Explaining the Neandertal admixture paradox via long-running but very infrequent admixture
This seems like an interesting theory that might explain two facts that do not appear to gel well with existing models of a single (or few) short and intense periods of admixture during Out-of-Africa. These are:
So, all in all, I'm fairly sympathetic to this model, and I'd be interested to see how it is received by experts in this field.
PLoS ONE 7(10): e47076. doi:10.1371/journal.pone.0047076
Extremely Rare Interbreeding Events Can Explain Neanderthal DNA in Living Humans
Armando G. M. Neves, Maurizio Serva
Considering the recent experimental discovery of Green et al that present-day non-Africans have 1 to 4% of their nuclear DNA of Neanderthal origin, we propose here a model which is able to quantify the genetic interbreeding between two subpopulations with equal fitness, living in the same geographic region. The model consists of a solvable system of deterministic ordinary differential equations containing as a stochastic ingredient a realization of the neutral Wright-Fisher process. By simulating the stochastic part of the model we are able to apply it to the interbreeding ofthe African ancestors of Eurasians and Middle Eastern Neanderthal subpopulations and estimate the only parameter of the model, which is the number of individuals per generation exchanged between subpopulations. Our results indicate that the amount of Neanderthal DNA in living non-Africans can be explained with maximum probability by the exchange of a single pair of individuals between the subpopulations at each 77 generations, but larger exchange frequencies are also allowed with sizeable probability. The results are compatible with a long coexistence time of 130,000 years, a total interbreeding population of order individuals, and with all living humans being descendants of Africans both for mitochondrial DNA and Y chromosome.
Link
- The complete absence of archaic Eurasian Y chromosomes and mtDNA in the modern human gene pool
- The fact that Neandertals did not appear to have gotten morphologically more different than modern humans over most of their late history, but rather the opposite, they became ever-more similar to modern humans
- it would be a very natural consequence of my favored scenario that modern humans (living probably in northern and eastern Africa) had Neandertal neighbors living in the Near East and southern Europe from which they were separated by geographical barriers, making admixture likely but not routine. The authors consider the idea that the admixture took place in the Near East, and even if some of it did not, I'd say most of it must have taken place there, since that is where maximal evidence of temporal co-existence between the two demes exists.
- it would not require an unlikely scenario of large-scale hybridization between very divergent demes: occasional gene flow could still occur, and could spread adaptations back and forth (explaining the phenotypic non-divergence), but the tendency of people to marry those like themselves (homogamy) would be preserved.
- It would be consistent with the re-writing of Out-of-Africa thanks to the halving of the autosomal rate. This would necessitate an early OoA and thus a longer occupation of parts of Asia by both sapiens and Neandertals.during which they may have occasionally interbred. So, not only were modern humans and Neandertals neighbors in Asia, but modern Eurasians are not descended from a fresh ~50ka Out-of-Sub-Saharan Africa expansion that would have rendered these long neighborly relations in the Near East irrelevant.
So, all in all, I'm fairly sympathetic to this model, and I'd be interested to see how it is received by experts in this field.
PLoS ONE 7(10): e47076. doi:10.1371/journal.pone.0047076
Extremely Rare Interbreeding Events Can Explain Neanderthal DNA in Living Humans
Armando G. M. Neves, Maurizio Serva
Considering the recent experimental discovery of Green et al that present-day non-Africans have 1 to 4% of their nuclear DNA of Neanderthal origin, we propose here a model which is able to quantify the genetic interbreeding between two subpopulations with equal fitness, living in the same geographic region. The model consists of a solvable system of deterministic ordinary differential equations containing as a stochastic ingredient a realization of the neutral Wright-Fisher process. By simulating the stochastic part of the model we are able to apply it to the interbreeding ofthe African ancestors of Eurasians and Middle Eastern Neanderthal subpopulations and estimate the only parameter of the model, which is the number of individuals per generation exchanged between subpopulations. Our results indicate that the amount of Neanderthal DNA in living non-Africans can be explained with maximum probability by the exchange of a single pair of individuals between the subpopulations at each 77 generations, but larger exchange frequencies are also allowed with sizeable probability. The results are compatible with a long coexistence time of 130,000 years, a total interbreeding population of order individuals, and with all living humans being descendants of Africans both for mitochondrial DNA and Y chromosome.
Link
October 23, 2012
Ancient European DNA assessment with 'globe10'
I had previously assessed the same using globe4. See post on globe10 and associated spreadsheet.
The results appear similar to previous analyses overall, with the main features being the presence of "Southern" in Neolithic farmers (which peaks in the Near East), and its absence in hunter-gatherers. Some of the "Amerindian"-like admixture that was evident in globe4 has been "absorbed" by the Atlantic_Baltic (main European) component, but it is interesting that the Swedish hunter-gatherers (Ajv52/Ajv70) continue to show some Amerindian as well as other eastern (Australasian/South Asian) admixture that is lacking in the other samples. These individuals are outside the range of modern populations, but they overall tend to map to the most similar Atlantic_Baltic component with the addition of some eastern influences.
Also of interest is the fact the Oetzi is the only sample which shows a slice of West Asian (5.7%) admixture in this analysis. This was also the case in the previous one using K7b (1.4%). Gok4, on the other hand, the fellow Neolithic individual from Sweden seems to lack this. The arrangement of the Big Three West Eurasian components (Southern/West Asian/Atlantic_Baltic) has subtly changed in this calculator, but it would be tempting, nonetheless, to see in the little West Asian admixture that Oetzi has but Gok4 and the Mesolithic samples seem to lack, something of the vanguard of the arrival of the West Asian component in Europe. Obviously more samples are needed, including ones from the most interesting regions of the Balkans and Anatolia.
The great human expansion (Henn et al. 2012)
I have been a rather outspoken critic of the "standard recent Out-of-Africa model" of human origins. A new paper by Henn, Cavalli-Sforza, and Feldman presents an up-to-date version of that model, and is quite useful as an overview of what I believe to be (and I'm sure the authors do not!) the passing paradigm.
From the paper:
From the paper:
If out species became behaviorally modern due to a population expansion circa 50ka, then we would expect different human populations to have split times of ~50ka. This is not, however, what we observe, but, rather, in all genetic systems (mtDNA, Y-chromosomes, and autosomal DNA), there is evidence for population splits within Homo sapiens of order 200ka, which were not, however, complete, but were followed by later episodes of admixture.
A recent paper estimated that the Khoe-San split from the rest of us ~100ka. Even if we disregard the use of a now-outdated mutation rate, this is still twice as old as the UP/LSA transition. The implication is clear, that at least in some part of our species, behavioral modernity c. 50ka did not spread through the spread of a new population, but through the spread of an idea. Now, let's flip this around, and go from South Africa to the Levant, where we do have evidence for a ~100ka split between a group of modern humans (the Mt. Carmel ones) and African humankind. If a population that split off ~100ka (and indeed, more likely 200 ka) within Africa is, nonetheless fully behaviorally modern by "cultural osmosis", so could the population of modern humans who lived in Asia pre-100ka: no need to invoke population replacement to explain the appearance of behavioral modernity.
The argument is simple: deep genetic population splits are no obstactle to the flow of culture in the case of Africa, so why postulate an obstacle to cultural flow (in whatever direction) between human groups with equal, or indeed much shallower split times?
I have written before about my distaste for Biblical-level bottlenecks, and here they are presented explicitly:
But, admixture matters. In the proximate sense, the diminution of genetic diversity from East Africa has never been established securely: to do so, one would need to isolate what is "diverse by admixture" and "diverse by antiquity". These proximate causes of increased diversity can be addressed because there are relatively unadmixed groups of people still in existence, and admixture LD has not had sufficient time to decay. This is particularly the case for many intermediate populations in the road Out-of-Africa, including East Africans, Near Eastern populations, South Asians, etc., all of which have evidence for recent admixture. Indeed, recent work has also established admixture within Africa itself, of both the recent and the archaic kind.
But, if the principle of admixture is accepted, then the possibility that it may have occurred in the distant past must also be entertained. In the absence of both LD-based evidence (which decays exponentially), and extant unadmixed populations (which tend to be absorbed or die out), older episodes of admixture will manifest themselves as little more than an excess of polymorphism, all the greater depending on the size of the introgressing element and its genetic divergence: a little admixture from a much diverged element will contribute a similar number of new alleles as a lot of admixture from a less diverged one.
In fact, we do see such an excess of polymorphism in Africans, and it will serve us well to remember that the Out-of-Africa bottleneck may have joined forces with an In-Africa-Admixture to create the contrast between African and Eurasian effective population sizes.
The story told in The great human expansion is, in my opinion, no longer believable. Three reasons have contributed to make it so:
PNAS doi: 10.1073/pnas.1212380109
The great human expansion
Brenna M. Henn et al.
Genetic and paleoanthropological evidence is in accord that today’s human population is the result of a great demic (demographic and geographic) expansion that began approximately 45,000 to 60,000 y ago in Africa and rapidly resulted in human occupation of almost all of the Earth’s habitable regions. Genomic data from contemporary humans suggest that this expansion was accompanied by a continuous loss of genetic diversity, a result of what is called the “serial founder effect.” In addition to genomic data, the serial founder effect model is now supported by the genetics of human parasites, morphology, and linguistics. This particular population history gave rise to the two defining features of genetic variation in humans: genomes from the substructured populations of Africa retain an exceptional number of unique variants, and there is a dramatic reduction in genetic diversity within populations living outside of Africa. These two patterns are relevant for medical genetic studies mapping genotypes to phenotypes and for inferring the power of natural selection in human history. It should be appreciated that the initial expansion and subsequent serial founder effect were determined by demographic and sociocultural factors associated with hunter-gatherer populations. How do we reconcile this major demic expansion with the population stability that followed for thousands years until the inventions of agriculture? We review advances in understanding the genetic diversity within Africa and the great human expansion out of Africa and offer hypotheses that can help to establish a more synthetic view of modern human evolution.
Link
From the paper:
Genetic data indicate that, approximately 45 to 60 kya, a very rapid population expansion occurred outside of Africa, and spread in all directions across the Eurasian continents, eventually populating the entire world.This is true. The question is whether this expansion originated in Africa itself, or in Eurasia, from people who had left Africa at a much earlier time. One aspect of this expansion that is often brought in defense of this hypothesis is the orderly diminution of genetic diversity outside Africa from the Near East. But, we ought to remember that "clines don't carry dates", and that particular one is consistent with an Out-of-Arabia dispersal of modern humans during the time in question.
From the paper:
However, current evidence indicates that this near-modern population did not persist in the Near East and was subsequently replaced by Neanderthals during the following glacial period, with little evidence of temporal overlap (5, 6). It is not until at least 50,000 y ago that evidence of behaviorally modern humans occurs in the archaeological record in the Near East.The evidence for behavioral modernity (the transition to the Upper Paleolithic and/or Lower Stone Age) appears near simultaneously around the planet and is thus no evidence for an Out-of-Africa event accompanying it.
If out species became behaviorally modern due to a population expansion circa 50ka, then we would expect different human populations to have split times of ~50ka. This is not, however, what we observe, but, rather, in all genetic systems (mtDNA, Y-chromosomes, and autosomal DNA), there is evidence for population splits within Homo sapiens of order 200ka, which were not, however, complete, but were followed by later episodes of admixture.
A recent paper estimated that the Khoe-San split from the rest of us ~100ka. Even if we disregard the use of a now-outdated mutation rate, this is still twice as old as the UP/LSA transition. The implication is clear, that at least in some part of our species, behavioral modernity c. 50ka did not spread through the spread of a new population, but through the spread of an idea. Now, let's flip this around, and go from South Africa to the Levant, where we do have evidence for a ~100ka split between a group of modern humans (the Mt. Carmel ones) and African humankind. If a population that split off ~100ka (and indeed, more likely 200 ka) within Africa is, nonetheless fully behaviorally modern by "cultural osmosis", so could the population of modern humans who lived in Asia pre-100ka: no need to invoke population replacement to explain the appearance of behavioral modernity.
The argument is simple: deep genetic population splits are no obstactle to the flow of culture in the case of Africa, so why postulate an obstacle to cultural flow (in whatever direction) between human groups with equal, or indeed much shallower split times?
I have written before about my distaste for Biblical-level bottlenecks, and here they are presented explicitly:
Resequencing studies have estimated the ancestral effective population size at 12,800 to 14,400, with a 5- to 10-fold bottleneck beginning approximately 65,000 to 50,000 y ago (although see ref. 15 for a bottleneck to only 450 individuals). It is generally assumed that the bottleneck occurred as a small group(s) with an effective population size of only approximately 1,000 to 2,500 individuals moved from the African continent into the Near East.This is of course, possible. But, the model writes the story, and if one assumes tree-like divergence of human populations sans admixture, then one will doubtlessly infer a story of migration going from the most diverse human populations, to the least diverse ones.
But, admixture matters. In the proximate sense, the diminution of genetic diversity from East Africa has never been established securely: to do so, one would need to isolate what is "diverse by admixture" and "diverse by antiquity". These proximate causes of increased diversity can be addressed because there are relatively unadmixed groups of people still in existence, and admixture LD has not had sufficient time to decay. This is particularly the case for many intermediate populations in the road Out-of-Africa, including East Africans, Near Eastern populations, South Asians, etc., all of which have evidence for recent admixture. Indeed, recent work has also established admixture within Africa itself, of both the recent and the archaic kind.
But, if the principle of admixture is accepted, then the possibility that it may have occurred in the distant past must also be entertained. In the absence of both LD-based evidence (which decays exponentially), and extant unadmixed populations (which tend to be absorbed or die out), older episodes of admixture will manifest themselves as little more than an excess of polymorphism, all the greater depending on the size of the introgressing element and its genetic divergence: a little admixture from a much diverged element will contribute a similar number of new alleles as a lot of admixture from a less diverged one.
In fact, we do see such an excess of polymorphism in Africans, and it will serve us well to remember that the Out-of-Africa bottleneck may have joined forces with an In-Africa-Admixture to create the contrast between African and Eurasian effective population sizes.
The story told in The great human expansion is, in my opinion, no longer believable. Three reasons have contributed to make it so:
- The publication of the Neandertal and Denisovan genomes have killed off any notion that the human tree blossomed in a vacuum, unperturbed by the other denizens of the Homo forest.
- Recalibration of the human autosomal mutation rate have revealed deep autosomal divergences within our species. These can be consistent with either (i) a recent expansion followed by admixture with divergent lineages, or (ii) old population structure within the species accompanied by cultural flow of behavioral modernity. I tend to support a mix of these ideas. What cannot have happened, however, is the model of a recent, simultaneous expansion responsible for both the spread of modern humans and behavioral modernity.
- While the ~60ka Out-of-Africans remain elusive, archaeologists have made steady progress in uncovering real links between Africa and Eurasia prior to 100ka. The Mousterian-using Mt. Carmel members of our species can no longer be discounted as the Out-of-Africa that failed, because they are now accompanied by Nubian Complex and Jebel Faya Arabians at around the same time.
PNAS doi: 10.1073/pnas.1212380109
The great human expansion
Brenna M. Henn et al.
Genetic and paleoanthropological evidence is in accord that today’s human population is the result of a great demic (demographic and geographic) expansion that began approximately 45,000 to 60,000 y ago in Africa and rapidly resulted in human occupation of almost all of the Earth’s habitable regions. Genomic data from contemporary humans suggest that this expansion was accompanied by a continuous loss of genetic diversity, a result of what is called the “serial founder effect.” In addition to genomic data, the serial founder effect model is now supported by the genetics of human parasites, morphology, and linguistics. This particular population history gave rise to the two defining features of genetic variation in humans: genomes from the substructured populations of Africa retain an exceptional number of unique variants, and there is a dramatic reduction in genetic diversity within populations living outside of Africa. These two patterns are relevant for medical genetic studies mapping genotypes to phenotypes and for inferring the power of natural selection in human history. It should be appreciated that the initial expansion and subsequent serial founder effect were determined by demographic and sociocultural factors associated with hunter-gatherer populations. How do we reconcile this major demic expansion with the population stability that followed for thousands years until the inventions of agriculture? We review advances in understanding the genetic diversity within Africa and the great human expansion out of Africa and offer hypotheses that can help to establish a more synthetic view of modern human evolution.
Link
October 22, 2012
Ancient mtDNA of first New Zealanders
PNAS doi: 10.1073/pnas.1209896109
Complete mitochondrial DNA genome sequences from the first New Zealanders
Michael Knapp et al.
The dispersal of modern humans across the globe began ∼65,000 y ago when people first left Africa and culminated with the settlement of East Polynesia, which occurred in the last 1,000 y. With the arrival of Polynesian canoes only 750 y ago, Aotearoa/New Zealand became the last major landmass to be permanently settled by humans. We present here complete mitochondrial genome sequences of the likely founding population of Aotearoa/New Zealand recovered from the archaeological site of Wairau Bar. These data represent complete mitochondrial genome sequences from ancient Polynesian voyagers and provide insights into the genetic diversity of human populations in the Pacific at the time of the settlement of East Polynesia.
Link
Complete mitochondrial DNA genome sequences from the first New Zealanders
Michael Knapp et al.
The dispersal of modern humans across the globe began ∼65,000 y ago when people first left Africa and culminated with the settlement of East Polynesia, which occurred in the last 1,000 y. With the arrival of Polynesian canoes only 750 y ago, Aotearoa/New Zealand became the last major landmass to be permanently settled by humans. We present here complete mitochondrial genome sequences of the likely founding population of Aotearoa/New Zealand recovered from the archaeological site of Wairau Bar. These data represent complete mitochondrial genome sequences from ancient Polynesian voyagers and provide insights into the genetic diversity of human populations in the Pacific at the time of the settlement of East Polynesia.
Link
October 21, 2012
Post-LGM expansion of mtDNA?
This issue keeps appearing and re-appearing. It is perhaps due to a tendency of conflating spatial population expansions with the proliferation of descendants within a lineage. The two are not necessarily related. Genetic-only methods can pick up on the signal of common descent and population growth, but cannot do the same for the signal of spatial expansion. Whether this growth happens (i) in situ for a long time, and only lately becomes a spatial expansion, or (ii) at the same time as the spatial expansion, or indeed (iii) long after it, will result in coalescences that precede, coincide with, or follow the actual spatial expansion event.
It is difficult to see how Europe was being filled up for thousands of years by a population taking advantage of post-glacial warming conditions, and, yet, when we actually look at ancient DNA from Europeans who lived just before the advent of farming, they show little evidence of possessing (m)any of the lineages that had been supposedly expanding in Europe since the LGM.
SCIENTIFIC REPORTS doi:10.1038/srep00745
MtDNA analysis of global populations support that major population expansions began before Neolithic Time
Hong-Xiang Zheng et al.
Agriculture resulted in extensive population growths and human activities. However, whether major human expansions started after Neolithic Time still remained controversial. With the benefit of 1000 Genome Project, we were able to analyze a total of 910 samples from 11 populations in Africa, Europe and Americas. From these random samples, we identified the expansion lineages and reconstructed the historical demographic variations. In all the three continents, we found that most major lineage expansions (11 out of 15 star lineages in Africa, all autochthonous lineages in Europe and America) coalesced before the first appearance of agriculture. Furthermore, major population expansions were estimated after Last Glacial Maximum but before Neolithic Time, also corresponding to the result of major lineage expansions. Considering results in current and previous study, global mtDNA evidence showed that rising temperature after Last Glacial Maximum offered amiable environments and might be the most important factor for prehistorical human expansions.
Link
It is difficult to see how Europe was being filled up for thousands of years by a population taking advantage of post-glacial warming conditions, and, yet, when we actually look at ancient DNA from Europeans who lived just before the advent of farming, they show little evidence of possessing (m)any of the lineages that had been supposedly expanding in Europe since the LGM.
SCIENTIFIC REPORTS doi:10.1038/srep00745
MtDNA analysis of global populations support that major population expansions began before Neolithic Time
Hong-Xiang Zheng et al.
Agriculture resulted in extensive population growths and human activities. However, whether major human expansions started after Neolithic Time still remained controversial. With the benefit of 1000 Genome Project, we were able to analyze a total of 910 samples from 11 populations in Africa, Europe and Americas. From these random samples, we identified the expansion lineages and reconstructed the historical demographic variations. In all the three continents, we found that most major lineage expansions (11 out of 15 star lineages in Africa, all autochthonous lineages in Europe and America) coalesced before the first appearance of agriculture. Furthermore, major population expansions were estimated after Last Glacial Maximum but before Neolithic Time, also corresponding to the result of major lineage expansions. Considering results in current and previous study, global mtDNA evidence showed that rising temperature after Last Glacial Maximum offered amiable environments and might be the most important factor for prehistorical human expansions.
Link
Ancient European DNA assessment with 'globe4'
In a previous experiment, I showed that ADMIXTURE at K=4 tracks the same signal of Amerindian-like admixture detected with f-statistics. I encapsulated that analysis in the globe4 calculator over at the Dodecad Project blog, and decided to use it to assess a few ancient European autosomal samples:
Please note that a very variable number of SNPs was extracted from these various samples. These results should be viewed as indicative of possible patterns that might be confirmed by a more thorough analysis. Also, please consult the globe4 post for more details on the methodology behind it, and the interpretation of the 4 components.
With these various caveats, I would say that these results seem to make some sense and to be fairly consistent with the scenario of Patterson et al. (2012):
- Oetzi and Gok4, the "farmers" seem to lack the Amerindian component
- Ajv52, and Ajv70, the northern hunter-gatherers seem to possess it
- Bra1, the Mesolithic Iberian seems to lack it as well
A broader context
The absence of the Amerindian-like admixture in South Indian Brahmins and Armenians, and its paucity Kurds and Iranians might indicate that this type of ancestry was not represented in ancient Armenians and Indo-Iranians. Indeed, all these populations possess less of this admixture than those of the North Caucasus. Cypriots possess none of it as well, where the Greek_D sample, a small 2.5% portion. In a previous analysis, I estimated a historical-era estimate of North European admixture in Greeks, and this admixture presumably incorporates the signal of Amerindian-like admixture. Additionally, an Iron Age individual from Bulgaria will soon be announced as being Sardinian-like.
The sum of these factors leads me to believe that the signal of Amerindian-like admixture did not play an important role in the formation of the Graeco-Phrygians (and their Armenian relatives) and the Indo-Iranians, or at least did so to an insignificant degree. As the former expanded westward from the PIE homeland, and the latter eastward, they would have had little opportunity to encounter this type of admixture; rather, they would have admixed with Sardinian-like individuals in the west, and Ancestral South Indian (ASI)-like or East Asian individuals in the east.
On the other hand, as Indo-European groups expanded into eastern Europe, setting off a chain of events that would eventually transform most of the northern part of the continent, and, in historical times, much of the rest of it, they would have met with Ajv-like individuals carrying the signal of Amerindian-like admixture, as well as the Oetzi/Sardinian-like farmers that had spread all the way to Scandinavia by the late Neolithic. The population formed by this mixture would have carried with it the signal of Amerindian-like ancestry, and would then transpose it across the continent. The signal would become increasingly muted westward and southward, and indeed this is what we observe.
UPDATE: It is interesting to see that South Indian Brahmins (both the Metspalu et al. sample, and my Iyer_D and Iyengar_D samples) lack this admixture, while Uttar Pradesh Brahmins do not, given the rolloff evidence for a more recent admixture of the latter. This is consistent with a historical admixture event, after the migration of Brahmin groups southwards, as described in that post.
October 19, 2012
Neandertals and modern humans may not have met in the southern Caucasus
There have been some recent indications that modern West Eurasians might not have precisely equal amounts of Neandertal ancestry, and that these differences may have been accented during prehistory. One possible explanation for this might be the fact that as modern humans expanded in Eurasia, they encountered different concentrations of Neandertals, and, in some places no Neandertals at all.
This hypothesis may be better resolved once a high coverage Neandertal genome is published, to complement the recent publication of the Denisova genome, as well as the new Altai Neandertals recently announced.
Journal of Human Evolution doi:dx.doi.org/10.1016/j.jhevol.2012.08.004
New chronology for the Middle Palaeolithic of the southern Caucasus suggests early demise of Neanderthals in this region
R. Pinhasi et al.
Neanderthal populations of the southern and northern Caucasus became locally extinct during the Late Pleistocene. The timing of their extinction is key to our understanding of the relationship between Neanderthals and anatomically modern humans (AMH) in Eurasia. Recent re-dating of the end of the Middle Palaeolithic (MP) at Mezmaiskaya Cave, northern Caucasus, and Ortvale Klde, southern Caucasus, suggests that Neanderthals did not survive after 39 ka cal BP (thousands of years ago, calibrated before present). Here we extend the analysis and present a revised regional chronology for MP occupational phases in western Georgia, based on a series of model-based Bayesian analyses of radiocarbon dated bone samples obtained from the caves of Sakajia, Ortvala and Bronze Cave. This allows the establishment of probability intervals for the onset and end of each of the dated levels and for the end of the MP occupation at the three sites.
Our results for Sakajia indicate that the end of the late Middle Palaeolithic (LMP) and start of the Upper Palaeolithic (UP) occurred between 40,200 and 37,140 cal BP. The end of the MP in the neighboring site of Ortvala occurred earlier at 43,540–41,420 cal BP (at 68.2% probability). The dating of MP layers from Bronze Cave confirms that it does not contain LMP phases.
These results imply that Neanderthals did not survive in the southern Caucasus after 37 ka cal BP, supporting a model of Neanderthal extinction around the same period as reported for the northern Caucasus and other regions of Europe. Taken together with previous reports of the earliest UP phases in the region and the lack of archaeological evidence for an in situ transition, these results indicate that AMH arrived in the Caucasus a few millennia after the Neanderthal demise and that the two species probably did not interact.
Link
This hypothesis may be better resolved once a high coverage Neandertal genome is published, to complement the recent publication of the Denisova genome, as well as the new Altai Neandertals recently announced.
Journal of Human Evolution doi:dx.doi.org/10.1016/j.jhevol.2012.08.004
New chronology for the Middle Palaeolithic of the southern Caucasus suggests early demise of Neanderthals in this region
R. Pinhasi et al.
Neanderthal populations of the southern and northern Caucasus became locally extinct during the Late Pleistocene. The timing of their extinction is key to our understanding of the relationship between Neanderthals and anatomically modern humans (AMH) in Eurasia. Recent re-dating of the end of the Middle Palaeolithic (MP) at Mezmaiskaya Cave, northern Caucasus, and Ortvale Klde, southern Caucasus, suggests that Neanderthals did not survive after 39 ka cal BP (thousands of years ago, calibrated before present). Here we extend the analysis and present a revised regional chronology for MP occupational phases in western Georgia, based on a series of model-based Bayesian analyses of radiocarbon dated bone samples obtained from the caves of Sakajia, Ortvala and Bronze Cave. This allows the establishment of probability intervals for the onset and end of each of the dated levels and for the end of the MP occupation at the three sites.
Our results for Sakajia indicate that the end of the late Middle Palaeolithic (LMP) and start of the Upper Palaeolithic (UP) occurred between 40,200 and 37,140 cal BP. The end of the MP in the neighboring site of Ortvala occurred earlier at 43,540–41,420 cal BP (at 68.2% probability). The dating of MP layers from Bronze Cave confirms that it does not contain LMP phases.
These results imply that Neanderthals did not survive in the southern Caucasus after 37 ka cal BP, supporting a model of Neanderthal extinction around the same period as reported for the northern Caucasus and other regions of Europe. Taken together with previous reports of the earliest UP phases in the region and the lack of archaeological evidence for an in situ transition, these results indicate that AMH arrived in the Caucasus a few millennia after the Neanderthal demise and that the two species probably did not interact.
Link
October 18, 2012
Neandertal-modern hybrid babies and their heads
A discussion in the comments of Gene Expression got me thinking about a potential scenario for modern-Neandertal interbreeding dynamics. That discussion dealt with difficulties in childbirth arising from population differences in birth canal/head size.
The main idea is simple, and I will rephrase it as follows: offspring of a big man and small woman will tend to have bigger heads relative to the size of the woman's birth canal. On the other hand, offspring of a small man and big woman will not have that problem.
We have some information about differences between Neandertals and modern humans. The former were shorter and more "lateral" skeletally, while UP modern humans appear to have been more linear and taller. Headwise, modern humans had more globular head shapes, while Neandertals more linear ones, with no major differences in brain size between the two species.
If the above are correct, then male Neandertal-female modern human pairings would have a potential problem. Birth is a complex process, but at the end of the day, the most important factor is probably whether the diameter of the head can "fit" in the birth canal: the more it does not fit, the more likely it would seem that a mishap for both mother and offspring would occur.
Combine elongated Neandertal heads with narrow modern human pelves, and you have a potential problem. I am not 100% sure that modern humans and Neandertals differed in pelvis shape, although it would be a reasonable consequence of their overall build, but the same pattern would occur if they did not, simply on account of their different head shapes.
An additional factor involves sexual dimorphism, since male babies tend to be larger than female ones, and so any problems associated with "parental mismatch" might be particularly troublesome for male births.
So, all in all, we have 4 different cases:
The main idea is simple, and I will rephrase it as follows: offspring of a big man and small woman will tend to have bigger heads relative to the size of the woman's birth canal. On the other hand, offspring of a small man and big woman will not have that problem.
We have some information about differences between Neandertals and modern humans. The former were shorter and more "lateral" skeletally, while UP modern humans appear to have been more linear and taller. Headwise, modern humans had more globular head shapes, while Neandertals more linear ones, with no major differences in brain size between the two species.
If the above are correct, then male Neandertal-female modern human pairings would have a potential problem. Birth is a complex process, but at the end of the day, the most important factor is probably whether the diameter of the head can "fit" in the birth canal: the more it does not fit, the more likely it would seem that a mishap for both mother and offspring would occur.
Combine elongated Neandertal heads with narrow modern human pelves, and you have a potential problem. I am not 100% sure that modern humans and Neandertals differed in pelvis shape, although it would be a reasonable consequence of their overall build, but the same pattern would occur if they did not, simply on account of their different head shapes.
An additional factor involves sexual dimorphism, since male babies tend to be larger than female ones, and so any problems associated with "parental mismatch" might be particularly troublesome for male births.
So, all in all, we have 4 different cases:
- Male H. n + Female H. s. => Male hybrid
- Male H. n + Female H. s. => Female hybrid
- Male H. s + Female H. n. => Male hybrid
- Male H. s + Female H. n. => Female hybrid
It would appear, on the basis of the preceding discussion, that 1-2 would be more troublesome than 3-4, and 1 most troublesome of all. On the other hand, 4 seems to be the most advantageous case.
The most interesting thing about modern-Neandertal admixture is that it seems to have left no traces in uniparental markers, and, indeed, the lack of mtDNA lineages of Neandertal origin has been used to argue against the plausibility of estimated Neandertal admixture percentages.
If my reasoning is correct, then case #4 is particularly worrying, since female hybrids with Neandertal mtDNA ought to be the most easy to bear, and would also be the ones who would contribute Neandertal mtDNA in a mixed population.
On the other hand, case #1 would explain the lack of Neandertal Y-chromosomes, since crossings between male Neandertals and female modern humans that produce male offspring might be particular troublesome, and they would also be the ones to introduce Neandertal Y-chromosomes in the population.
Of course, we don't know enough about the dynamics of the admixture process; it might be possible that other factors influence the abundance of the four cases, both biological and cultural. For example, if modern humans had a behavioral advantage, then modern males may contribute most admixture, and this would make the worrying case #4 even more difficult to explain. On the other hand, how did the admixture take place? bride-stealing vs. rape would result in potential offspring being raised in different groups (father's vs. mother's), and there may also have been unknown cultural taboos involving admixture and offpsring produced from it.
In any case, this brief excursus may be useful for anyone thinking of writing some palaeo-fiction set in the Upper Paleolithic, and I'd love to hear from people who have data at hand that might be pertinent to the above discussion.
rolloff analysis of Lezgins as Sardinian+Burusho
I have carried out rolloff analysis of Lezgins, a Northeast Caucasian population that is of particular interest due to it being modal for the "Dagestan" component whose long-distance relationships with Western Europe and South Asia have triggered a great deal of followup investigation on my part.
The Lezgins are also interesting for other reasons: they may be one of the populations related to the Kura-Araxes culture; they possess a high frequency of Y-haplogroup R1b, so they may be related to the migration that brought this haplogroup into Europe from West Asia.
In my previous analysis of the French using the same reference populations, I speculated that their signal of admixture may involve admixture between a Sardinian-like and a West Asian population in Asia itself circa 7,000 years ago, followed by a later expansion into Europe. And, in my analysis of Lithuanians and Ukrainians, I discovered a somewhat less "old" signal of admixture involving South Asian+North European references with a mean value of 5.5-6.3ky for the various population pairs.
The exponential fit for the Lezgins can be seen below:
The admixture time estimate is 198.773 +/- 70.649 generations or 5,760 +/- 2,050 years. This is not very precise, but seems consistent with the two phenomena described above. It also seems to contrast with the much younger signal for Armenians.
The Lezgins are also interesting for other reasons: they may be one of the populations related to the Kura-Araxes culture; they possess a high frequency of Y-haplogroup R1b, so they may be related to the migration that brought this haplogroup into Europe from West Asia.
In my previous analysis of the French using the same reference populations, I speculated that their signal of admixture may involve admixture between a Sardinian-like and a West Asian population in Asia itself circa 7,000 years ago, followed by a later expansion into Europe. And, in my analysis of Lithuanians and Ukrainians, I discovered a somewhat less "old" signal of admixture involving South Asian+North European references with a mean value of 5.5-6.3ky for the various population pairs.
The exponential fit for the Lezgins can be seen below:
The admixture time estimate is 198.773 +/- 70.649 generations or 5,760 +/- 2,050 years. This is not very precise, but seems consistent with the two phenomena described above. It also seems to contrast with the much younger signal for Armenians.
Relatives/duplicates in ADMIXTURE
The presence of relatives in a dataset tends to throw ADMIXTURE out, but this does not always happen. In particular, I've noticed that at low K, relatives do not appear to form their own hyper-specific clusters. A good example of this is the Yunusbayev et al. Armenians_Y sample (N=16) that happens to include what appears to be a common individual (or a twin?) with my own own Armenian_D sample from the Dodecad Project. This was discovered the last time I ran ADMIXTURE, so I henceforth began using a subset of 15 Armenians (Armenians_15_Y) from that dataset whenever I also included my Dodecad sample.
In my current ongoing analysis of the world dataset, I included two versions of the Sakilli, Paniya, and Malayan samples, from Behar et al. and Chaubey et al. I believe that HarrappaDNA Project has previously identified that some of these are not exactly the same individuals, so I wanted to see what the ancestry of all these individuals was, to help me decide which ones to keep.
Here are the K=5 ancestral proportions of the Behar et al. Sakilli:
GSM536813 10.2 7.8 2.2 0 79.9
GSM536814 8.5 9.3 2.1 0 80.0
GSM536815 9.7 7.9 3.6 0 78.8
GSM536816 8.8 8.7 2.1 0 80.4
In my current ongoing analysis of the world dataset, I included two versions of the Sakilli, Paniya, and Malayan samples, from Behar et al. and Chaubey et al. I believe that HarrappaDNA Project has previously identified that some of these are not exactly the same individuals, so I wanted to see what the ancestry of all these individuals was, to help me decide which ones to keep.
Here are the K=5 ancestral proportions of the Behar et al. Sakilli:
GSM536813 10.2 7.8 2.2 0 79.9
GSM536814 8.5 9.3 2.1 0 80.0
GSM536815 9.7 7.9 3.6 0 78.8
GSM536816 8.8 8.7 2.1 0 80.4
and of the Chaubey et al. Sakilli:
SAKD60 10.2 7.8 2.2 0 79.9
SAKD72 9.7 7.9 3.6 0 78.8
SAKD75 8.8 8.7 2.1 0 80.4
SAKD64 8.5 9.4 2.1 0 80.0
These appear to be the same individuals, which was confirmed by IBD analysis.
The Malayan individuals also appear to be the same:
GSM536915 0.3 15.5 2.7 0 81.6
GSM536812 3.3 16.6 2.8 0 77.3
A382 0.3 15.5 2.7 0 81.6
MLYA383 3.3 16.6 2.8 0 77.3
But, as noticed by HAP, the Paniya individuals are not the same:
GSM536916 5.1 11.2 2.2 0.0 81.6
GSM536806 0.4 69.7 0.0 4.3 25.6
GSM536807 0.0 79.7 0.0 2.4 18.0
GSM536808 0.0 77.5 0.5 1.7 20.3
2953 D36 5.1 11.2 2.2 0.0 81.6
2954 PNYD9 0.0 19.8 2.5 0.6 77.1
2955 PNYD3 0.0 21.2 1.5 0.0 77.3
2956 PNYD1 0.0 21.7 2.7 0.3 75.2
As I move forward in my "world" analysis, I've decided to drop GSM536916 and the Chaubey et al. versions of Sakilli and Malayan. Thus, PANIYA will refer to the Southeast Asian-like individuals of the Behar et al. set, and Paniya_Ch to the South Asian-like individuals of the Chaubey et al. set, with one copy of the duplicated individual removed.
ADMIXTURE tracks Amerindian-like admixture in northern Europe
I have recently assembled a new "world" dataset of 4,280 individuals that I am currently incrementally analyzing with ADMIXTURE. But, I noticed an interesting pattern at K=4 that I wanted to share right away.
4 ancestral populations emerge at this level of resolution, which I have named: European, Asian, African, Amerindian. The names aren't important, and you can replace them with whatever you prefer.
The interesting thing about this K=4 analysis is that European populations show evidence of Amerindian admixture, consistent with the pattern inferred using f-statistics, where European populations show admixture between Sardinians and a Karitiana-like population.
This pattern may have emerged at previous ADMIXTURE analyses at this level of resolution, but thanks to the f3 evidence presented in previous posts, it is now clear that it is no quirk of ADMIXTURE, but indicative of a real (albeit still rather mysterious) pattern of gene flow that differentially affected European populations.
For example, the Irish_D population has 7.6% of the Amerindian component, and so do HGDP Orcadians. HGDP Sardinians have only 1.7% of it, which appears to be the minimum in Europe, with French_Basque having more at 4.6%.
Another interesting observation is that West Eurasian populations that show an excess of East Eurasian-like admixture appear to be doing so for two separate reasons. For example, HGDP Russians have 11.7% of Amerindian component, but also 4.5% of "Asian", and 1000 Genomes Finns have 3.3% Asian and 12% Amerindian. Behar et al. (2010) Turks, on the other hand, have 9.9% Asian and 2.2% Amerindian. All these populations are East Eurasian-shifted relative to Sardinians, a pattern which can also be observed by looking at the K=3 analysis, but for apparently different reasons.
The pattern for Near Eastern populations is also interesting. For example, Yunusbayev et al. (2011) Armenians have 0% of the Amerindian component, and 5.7% of the Asian, and all three HGDP Arab populations (Druze, Palestinian, Bedouin) also have 0% of the Amerindian component, with variable levels of the Asian.
It would appear that whatever process contributed Amerindian-like admixture in Europeans, minimally affected Near Eastern populations, with Sardinians being demonstrably related to Neolithic Europeans (thanks to ancient DNA evidence), tilting towards the Near Eastern pattern. On the other hand, Near Eastern populations show evidence of Asian admixture, which probably involves unresolved East Asian/ASI ancestry, and will be resolved at higher K. Sardinians appear to be at the end of three clines: (i) Amerindian-like cline of Europe-Siberia-Americas, (ii) East Asian-like cline of Europe-Central Asia/Siberia-East Asia, (iii) ASI-like cline of Europe-Near East-South Asia. These are separate, but not independent phenomena.
To confirm that the signal picked up by ADMIXTURE tracks the signal picked up by ADMIXTOOLS formal tests, I calculated the following D-statistic:
where European is any population with a sample size of at least 10, and which belonged at 99% in the European+Amerindian components:
And, here is a scatterplot:
The correlation is clear, and the Pearson coefficient is -0.96. This means that populations with higher % Amerindian, as estimated by ADMIXTURE, also show higher D-statistic evidence for admixture.
What of the actual estimates of admixture produced by ADMIXTURE? Using the F4 ratio test, I recently showed that African admixture in Sardinians confounds estimates of Amerindian-like admixture in northern Europeans and vice versa (Amerindian-like admixture in northern Europeans confounds African admixture in Sardinians).
In that experiment, I "scrubbed" Sardinians to remove segments of African ancestry, and showed that estimates of Amerindian-like admixture in the CEU population diminished from 13.9% to 8.8%. The latter seems reasonably close to the 7.1% inferred by ADMIXTURE.
On balance, I would say that ADMIXTURE at K=4 provides a good proxy for the effect described in Patterson et al. (2012). Its results are more difficult to interpret, because its underlying model does not take into account evolutionary relationships between populations. On the other hand, it has the advantage of being able to handle multiple ancestral populations, and has consistently proven able to generate useful data that correlate well with those from other techniques of population genetics.
4 ancestral populations emerge at this level of resolution, which I have named: European, Asian, African, Amerindian. The names aren't important, and you can replace them with whatever you prefer.
The interesting thing about this K=4 analysis is that European populations show evidence of Amerindian admixture, consistent with the pattern inferred using f-statistics, where European populations show admixture between Sardinians and a Karitiana-like population.
This pattern may have emerged at previous ADMIXTURE analyses at this level of resolution, but thanks to the f3 evidence presented in previous posts, it is now clear that it is no quirk of ADMIXTURE, but indicative of a real (albeit still rather mysterious) pattern of gene flow that differentially affected European populations.
For example, the Irish_D population has 7.6% of the Amerindian component, and so do HGDP Orcadians. HGDP Sardinians have only 1.7% of it, which appears to be the minimum in Europe, with French_Basque having more at 4.6%.
Another interesting observation is that West Eurasian populations that show an excess of East Eurasian-like admixture appear to be doing so for two separate reasons. For example, HGDP Russians have 11.7% of Amerindian component, but also 4.5% of "Asian", and 1000 Genomes Finns have 3.3% Asian and 12% Amerindian. Behar et al. (2010) Turks, on the other hand, have 9.9% Asian and 2.2% Amerindian. All these populations are East Eurasian-shifted relative to Sardinians, a pattern which can also be observed by looking at the K=3 analysis, but for apparently different reasons.
The pattern for Near Eastern populations is also interesting. For example, Yunusbayev et al. (2011) Armenians have 0% of the Amerindian component, and 5.7% of the Asian, and all three HGDP Arab populations (Druze, Palestinian, Bedouin) also have 0% of the Amerindian component, with variable levels of the Asian.
It would appear that whatever process contributed Amerindian-like admixture in Europeans, minimally affected Near Eastern populations, with Sardinians being demonstrably related to Neolithic Europeans (thanks to ancient DNA evidence), tilting towards the Near Eastern pattern. On the other hand, Near Eastern populations show evidence of Asian admixture, which probably involves unresolved East Asian/ASI ancestry, and will be resolved at higher K. Sardinians appear to be at the end of three clines: (i) Amerindian-like cline of Europe-Siberia-Americas, (ii) East Asian-like cline of Europe-Central Asia/Siberia-East Asia, (iii) ASI-like cline of Europe-Near East-South Asia. These are separate, but not independent phenomena.
To confirm that the signal picked up by ADMIXTURE tracks the signal picked up by ADMIXTOOLS formal tests, I calculated the following D-statistic:
D(Sardinian, European, Karitiana, San)
where European is any population with a sample size of at least 10, and which belonged at 99% in the European+Amerindian components:
And, here is a scatterplot:
The correlation is clear, and the Pearson coefficient is -0.96. This means that populations with higher % Amerindian, as estimated by ADMIXTURE, also show higher D-statistic evidence for admixture.
What of the actual estimates of admixture produced by ADMIXTURE? Using the F4 ratio test, I recently showed that African admixture in Sardinians confounds estimates of Amerindian-like admixture in northern Europeans and vice versa (Amerindian-like admixture in northern Europeans confounds African admixture in Sardinians).
In that experiment, I "scrubbed" Sardinians to remove segments of African ancestry, and showed that estimates of Amerindian-like admixture in the CEU population diminished from 13.9% to 8.8%. The latter seems reasonably close to the 7.1% inferred by ADMIXTURE.
On balance, I would say that ADMIXTURE at K=4 provides a good proxy for the effect described in Patterson et al. (2012). Its results are more difficult to interpret, because its underlying model does not take into account evolutionary relationships between populations. On the other hand, it has the advantage of being able to handle multiple ancestral populations, and has consistently proven able to generate useful data that correlate well with those from other techniques of population genetics.
Neandertals in North Africa
Let me enter the following points which might be very relevant to the finding that in North Africa: "the Neandertal's genetic signal is higher in populations with a local, pre-Neolithic North African ancestry".
First of all, this is unexpected if Neandertal admixture took place in the Near East; if that were the case, then Near Eastern back-migrants would be more Neandertal-like than aboriginal Homo sapiens that had not participated in the Out-of-Africa event.
Second, I have followed up on John Hawks' suggestion that UP Europeans were more Neandertal-admixed than current Europeans, and using Oetzi's genome, discovered that potentially this is true. This is also unexpected if admixture with Neandertals took place in the Near East.
A link between aboriginal North Africans and UP Europeans of course exists: relationships between the Mechta-Afalou and Cro-Magnoids have long been recognized in physical anthropology.
An even more remote link involves Jebel Irhoud 1, the first modern human whose skull we possess from North Africa. Not only were the associated industries Mousterian (same as European Neandertals), but the skull itself was originally considered to be an African Neandertal, before it was reclassified as a member of H. sapiens.
I will update this entry after reading the paper with any further observations.
PLoS ONE 7(10): e47765. doi:10.1371/journal.pone.0047765
North African Populations Carry the Signature of Admixture with Neandertals
Federico Sánchez-Quinto et al.
One of the main findings derived from the analysis of the Neandertal genome was the evidence for admixture between Neandertals and non-African modern humans. An alternative scenario is that the ancestral population of non-Africans was closer to Neandertals than to Africans because of ancient population substructure. Thus, the study of North African populations is crucial for testing both hypotheses. We analyzed a total of 780,000 SNPs in 125 individuals representing seven different North African locations and searched for their ancestral/derived state in comparison to different human populations and Neandertals. We found that North African populations have a significant excess of derived alleles shared with Neandertals, when compared to sub-Saharan Africans. This excess is similar to that found in non-African humans, a fact that can be interpreted as a sign of Neandertal admixture. Furthermore, the Neandertal's genetic signal is higher in populations with a local, pre-Neolithic North African ancestry. Therefore, the detected ancient admixture is not due to recent Near Eastern or European migrations. Sub-Saharan populations are the only ones not affected by the admixture event with Neandertals.
Link
First of all, this is unexpected if Neandertal admixture took place in the Near East; if that were the case, then Near Eastern back-migrants would be more Neandertal-like than aboriginal Homo sapiens that had not participated in the Out-of-Africa event.
Second, I have followed up on John Hawks' suggestion that UP Europeans were more Neandertal-admixed than current Europeans, and using Oetzi's genome, discovered that potentially this is true. This is also unexpected if admixture with Neandertals took place in the Near East.
A link between aboriginal North Africans and UP Europeans of course exists: relationships between the Mechta-Afalou and Cro-Magnoids have long been recognized in physical anthropology.
An even more remote link involves Jebel Irhoud 1, the first modern human whose skull we possess from North Africa. Not only were the associated industries Mousterian (same as European Neandertals), but the skull itself was originally considered to be an African Neandertal, before it was reclassified as a member of H. sapiens.
I will update this entry after reading the paper with any further observations.
PLoS ONE 7(10): e47765. doi:10.1371/journal.pone.0047765
North African Populations Carry the Signature of Admixture with Neandertals
Federico Sánchez-Quinto et al.
One of the main findings derived from the analysis of the Neandertal genome was the evidence for admixture between Neandertals and non-African modern humans. An alternative scenario is that the ancestral population of non-Africans was closer to Neandertals than to Africans because of ancient population substructure. Thus, the study of North African populations is crucial for testing both hypotheses. We analyzed a total of 780,000 SNPs in 125 individuals representing seven different North African locations and searched for their ancestral/derived state in comparison to different human populations and Neandertals. We found that North African populations have a significant excess of derived alleles shared with Neandertals, when compared to sub-Saharan Africans. This excess is similar to that found in non-African humans, a fact that can be interpreted as a sign of Neandertal admixture. Furthermore, the Neandertal's genetic signal is higher in populations with a local, pre-Neolithic North African ancestry. Therefore, the detected ancient admixture is not due to recent Near Eastern or European migrations. Sub-Saharan populations are the only ones not affected by the admixture event with Neandertals.
Link
October 17, 2012
A nuanced reading of Earnest Hooton
AJPA DOI: 10.1002/ajpa.22162
Two faces of Earnest A. Hooton
Eugene Giles
The American Anthropological Association's multimedia project, “Race: Are We So Different?” alleges that Earnest A. Hooton (1887–1954) of Harvard University was a racist eugenicist who “perhaps more than any other scientist of his time… did more to establish racial stereotypes…” and infers racism from his having sat on a National Research Council Committee on the Negro in the 1920s. I take issue with this perspective to argue against Hooton as a racist by exploring Hooton's relationship with African American students, particularly Caroline Bond Day, and with the National Association for the Advancement of Colored People when it awarded a medal to Charles R. Drew, M.D. In the heyday of eugenics, Hooton was an atypical eugenicist in espousing a resolutely nonracial view of the woes of humankind perpetuated by what he considered the biologically unfit. As eugenics and Nazism became conflated in the late 1930s, Hooton hewed to a path that was more antiracist than many of his anthropological colleagues and publicly disputed Nazi racial ideology. Am J Phys Anthropol 2012. © 2012 Wiley Periodicals, Inc.
Link
Two faces of Earnest A. Hooton
Eugene Giles
The American Anthropological Association's multimedia project, “Race: Are We So Different?” alleges that Earnest A. Hooton (1887–1954) of Harvard University was a racist eugenicist who “perhaps more than any other scientist of his time… did more to establish racial stereotypes…” and infers racism from his having sat on a National Research Council Committee on the Negro in the 1920s. I take issue with this perspective to argue against Hooton as a racist by exploring Hooton's relationship with African American students, particularly Caroline Bond Day, and with the National Association for the Advancement of Colored People when it awarded a medal to Charles R. Drew, M.D. In the heyday of eugenics, Hooton was an atypical eugenicist in espousing a resolutely nonracial view of the woes of humankind perpetuated by what he considered the biologically unfit. As eugenics and Nazism became conflated in the late 1930s, Hooton hewed to a path that was more antiracist than many of his anthropological colleagues and publicly disputed Nazi racial ideology. Am J Phys Anthropol 2012. © 2012 Wiley Periodicals, Inc.
Link
Ancient mtDNA haplogroup X2 from Central Europe
Davidski reminds me of a paper by Lee et al. I had posted the abstract of, but did not comment on. He highlights the fact that mtDNA haplogroup X2 has been detected at this site (3.6-2.8ky cal BC) but not in earlier LBK Neolithic Europeans. Furthermore, he attributes the arrival of X2 in Europe to "Northwest Eurasians":
Of related interest, mtDNA haplogroup X2b has been detected in Iron Age "princely burials" from the same location and by the same group. Also from Reidla et al.:
Therefore, we can probably reject Davidski's speculation...
Reading the quotes below, I can’t help thinking that X2 lineages in Europe might be associated with the arrival of the so called Northwest Eurasians of North/Central/East Europe and the North Caucasus, while X1 with the earlier migrations of the Sardinian-like Southwest Eurasians of Mediterranean Europe, North Africa and the Near East.
However, mtDNA haplogroup X2 seems to have originated in the Near East:
Finally, phylogeography of the subclades of haplogroup X suggests that the Near East is the likely geographical source for the spread of subhaplogroup X2, and the associated population dispersal occurred around, or after, the LGM when the climate ameliorated. The presence of a daughter clade in northern Native Americans testifies to the range of this population expansion.Moreover, it occurs at a higher frequency in Southern Europeans than Northern Europeans and is well-represented in the Caucasus, Near East, and even Africa. These twin facts are inconsistent with it being related to "Northwest Eurasians", however that hypothetical people is defined.
Of related interest, mtDNA haplogroup X2b has been detected in Iron Age "princely burials" from the same location and by the same group. Also from Reidla et al.:
The sister groups X2b and X2c (X1 and X2, respectively, in the work of Herrnstadt et al. 2002) encompass one-third of the European sequences (excluding the samples from the North Caucasus). It is of interest that some North African sequences (from Morocco and Algeria) belong to X2b as well. Subhaplogroup X2b shows a diversity that is consistent with a postglacial population expansion in both West Eurasia and North Africa.Fernandes et al. (2012) consider X2b to be of European origin. X2 has been discovered in a Megalithic long mound from France (4.2ky cal BP), and in abundance at Treilles (c. 3,000 BC), in the latter case associated with a predominantly Y-haplogroup G2a (with some I-P37.2) population. In Jean Manco's excellent compendium, X2b is also listed as being present in Neolithic Portugal (3,400 years BC), and X2j in Neolithic Germany (4625-4250 BC); the latter is said to be "North African" by Fernandes et al. (2012).
Therefore, we can probably reject Davidski's speculation...
So, X2 has been located at multiple late Neolithic sites in Central Europe, including the Corded Ware burial ground at Eulau, Eastern Germany. Of course, that’s also where Y-chromosome haplogroup R1a was found (see here). I suspect this wasn’t a coincidence and it’s likely these markers entered Europe together from the east, probably between 4,000 and 3,000 B.C.
X2 shows no association with northern Europeans at present, and occurs in ancient DNA samples from Western Europe that show no indication of being related to Y-haplogroup R1a at all, and even precede the hypothetical 4-3ky BC entry window.
Also of interest is that no X2 was mentioned in recent published data from Ukraine and West Siberia, and none of it was detected in Mesolithic Europeans. So, it seems that X2 variants entered Europe during the Neolithic, and there is no indication that they did so with Davidski's hypothetical R1a-bearing Northwest Europeans.
Also of interest is that no X2 was mentioned in recent published data from Ukraine and West Siberia, and none of it was detected in Mesolithic Europeans. So, it seems that X2 variants entered Europe during the Neolithic, and there is no indication that they did so with Davidski's hypothetical R1a-bearing Northwest Europeans.
The tangled web of humanity
Indian populations are composed of two ancestral components: Ancestral North Indians (ANI) and Ancestral South Indians (ASI), discovered by Reich et al. (2009). In that paper, it was also shown that ASI forms a clade with East Eurasians, while ANI does so with West Eurasians.
Consider the following D-statistic:
As we shall see, this takes positive values, consistent with the idea of gene flow between Europeans and Indians at the exclusion of Sardinians. However, this gene flow may involve either the West Eurasian component in the ancestry of Indians (i.e., this component is more related to Europeans than to Sardinians), or to the ASI component (which is related to Europeans via the common "red arrow" portions of ancestry).
We can figure out what is going on by trying different Indian populations along the Indian Cline, and seeing whether the D-statistic is inflated/deflated in populations of greater ANI/ASI ancestry.
Here are the results:
Russian Orcadian French Lithuanians ANI
Mala 0.0153 0.0120 0.0088 0.0131 38.86
Madiga 0.0153 0.0122 0.0091 0.0111 40.66
Chenchu 0.0157 0.0108 0.0088 0.0115 40.76
Bhil 0.0149 0.0115 0.0086 0.0124 42.96
Satnami 0.0166 0.0125 0.0091 0.0126 43.06
Kurumba 0.0156 0.0117 0.0095 0.0121 43.26
Kamsali 0.0139 0.0105 0.0088 0.0098 44.56
Vysya 0.0130 0.0099 0.0083 0.0102 46.26
Lodi 0.0143 0.0124 0.0092 0.0125 49.96
Naidu 0.0138 0.0104 0.0092 0.0108 50.16
Tharu 0.0150 0.0112 0.0095 0.0118 51.06
Velama 0.0126 0.0107 0.0083 0.0095 54.76
Srivastava 0.0144 0.0124 0.0091 0.0116 56.46
Meghawal 0.0131 0.0107 0.0088 0.0117 60.36
Vaish 0.0143 0.0144 0.0099 0.0128 62.66
Kashmiri_Pandit 0.0119 0.0116 0.0090 0.0116 70.66
Sindhi 0.0106 0.0112 0.0095 0.0111 73.76
Pathan 0.0098 0.0114 0.0087 0.0106 76.96
In other words, the evidence for gene flow between Russians and Indians is maximized when south Indian (ASI-rich) populations are used.
Patterson et al. (2012) published a different pattern: non-Sardinian Europeans have North Eurasian-like ancestry that links them to Amerindian populations. It is thus possible that ASI and the East Eurasian-like admixture in North Europeans may share a common evolutionary history:
Now, consider a hypothetical population of the Indian Cline. A European population is related to it both via its ANI/West Eurasian ancestry, but also via its ASI ancestry, because the East_Eurasian component in Europeans shares a portion of ancestry (indicated by the red arrow) with ASI.
Sardinians lack (or have less of) this "red arrow" portion of ancestry.
It is also possible that ANI itself may have some East_Eurasian ancestry, like Europeans do; this is not indicated in the figure. More on this later.
D(European, Sardinian, Indian, San)
As we shall see, this takes positive values, consistent with the idea of gene flow between Europeans and Indians at the exclusion of Sardinians. However, this gene flow may involve either the West Eurasian component in the ancestry of Indians (i.e., this component is more related to Europeans than to Sardinians), or to the ASI component (which is related to Europeans via the common "red arrow" portions of ancestry).
We can figure out what is going on by trying different Indian populations along the Indian Cline, and seeing whether the D-statistic is inflated/deflated in populations of greater ANI/ASI ancestry.
Here are the results:
Russian Orcadian French Lithuanians ANI
Mala 0.0153 0.0120 0.0088 0.0131 38.86
Madiga 0.0153 0.0122 0.0091 0.0111 40.66
Chenchu 0.0157 0.0108 0.0088 0.0115 40.76
Bhil 0.0149 0.0115 0.0086 0.0124 42.96
Satnami 0.0166 0.0125 0.0091 0.0126 43.06
Kurumba 0.0156 0.0117 0.0095 0.0121 43.26
Kamsali 0.0139 0.0105 0.0088 0.0098 44.56
Vysya 0.0130 0.0099 0.0083 0.0102 46.26
Lodi 0.0143 0.0124 0.0092 0.0125 49.96
Naidu 0.0138 0.0104 0.0092 0.0108 50.16
Tharu 0.0150 0.0112 0.0095 0.0118 51.06
Velama 0.0126 0.0107 0.0083 0.0095 54.76
Srivastava 0.0144 0.0124 0.0091 0.0116 56.46
Meghawal 0.0131 0.0107 0.0088 0.0117 60.36
Vaish 0.0143 0.0144 0.0099 0.0128 62.66
Kashmiri_Pandit 0.0119 0.0116 0.0090 0.0116 70.66
Sindhi 0.0106 0.0112 0.0095 0.0111 73.76
Pathan 0.0098 0.0114 0.0087 0.0106 76.96
For each Indian Cline population, I list the ANI percentage, as estimated by Reich et al. (2009) in the last column, and the D-statistic of the above given form for different pairs of Indian and European populations.
We can plot the D-statistic vs. ANI for each of our European populations:
The correlation coefficients confirm the visual impression, that for the HGDP Russians there is a significantly negative relationship between ANI admixture in an Indian Cline population and the D-statistic:
Russian Orcadian French Lithuanians
-0.8631118 0.08670188 0.1870127 -0.1889908
In other words, the evidence for gene flow between Russians and Indians is maximized when south Indian (ASI-rich) populations are used.
The lack of a clear pattern in the other three populations is itself interesting. One possible explanation involves East Eurasian-like admixture in the ANI, a conjecture which would make sense, given that all non-Sardinian continental West Eurasians seem to possess it.
If that is true, then as we go "south" along the Indian Cline, ASI related admixture inflates the D-statistic by increasing the "red arrow" overlap with the East Eurasian-like admixture in Europeans. As we go "north" along this cline, then the D-statistic decreases, due to ASI-reduction, but also increases, due to East Eurasian-like admixture in ANI, with an end result of no clear pattern in the superposition of processes.
In any case, this is an interesting example of a crisscrossing type of admixture where unrelated processes (east Eurasian-like admixture in Russians and ASI admixture in Indians) combine to present an unusual effect.
If that is true, then as we go "south" along the Indian Cline, ASI related admixture inflates the D-statistic by increasing the "red arrow" overlap with the East Eurasian-like admixture in Europeans. As we go "north" along this cline, then the D-statistic decreases, due to ASI-reduction, but also increases, due to East Eurasian-like admixture in ANI, with an end result of no clear pattern in the superposition of processes.
In any case, this is an interesting example of a crisscrossing type of admixture where unrelated processes (east Eurasian-like admixture in Russians and ASI admixture in Indians) combine to present an unusual effect.
October 16, 2012
Compasses would have pointed south for 440 years ~41 thousand years ago.
Recent research indicates that when the Campanian Ignimbrite event occurred, the Neandertals were already on the way out. I'd say that the circa 40ka period would make the ideal setting for some good palaeo-fiction. You have volcanic explosions, modern humans replacing Neandertals, magnetic field reversals, and a new set of characters in the mysterious Denisovans. This stuff practically writes itself. On that topic, does anyone have any good prehistoric fiction recommendations?
Earth and Planetary Science Letters Volumes 351–352, 15 October 2012, Pages 54–69
Dynamics of the Laschamp geomagnetic excursion from Black Sea sediments
N.R. Nowaczyk et al.
Investigated sediment cores from the southeastern Black Sea provide a high-resolution record from mid latitudes of the Laschamp geomagnetic polarity excursion. Age constraints are provided by 16 AMS 14C ages, identification of the Campanian Ignimbrite tephra (39.28±0.11 ka), and by detailed tuning of sedimentologic parameters of the Black Sea sediments to the oxygen isotope record from the Greenland NGRIP ice core. According to the derived age model, virtual geomagnetic pole (VGP) positions during the Laschamp excursion persisted in Antarctica for an estimated 440 yr, making the Laschamp excursion a short-lived event with fully reversed polarity directions. The reversed phase, centred at 41.0 ka, is associated with a significant field intensity recovery to 20% of the preceding strong field maximum at ∼50 ka. Recorded field reversals of the Laschamp excursion, lasting only an estimated ∼250 yr, are characterized by low relative paleointensities (5% relative to 50 ka). The central, fully reversed phase of the Laschamp excursion is bracketed by VGP excursions to the Sargasso Sea (∼41.9 ka) and to the Labrador Sea (∼39.6 ka). Paleomagnetic results from the Black Sea are in excellent agreement with VGP data from the French type locality which facilitates the chronological ordering of the non-superposed lavas that crop out at Laschamp–Olby. In addition, VGPs between 34 and 35 ka reach low northerly to equatorial latitudes during a clockwise loop, inferred to be the Mono lake excursion.
Link
Earth and Planetary Science Letters Volumes 351–352, 15 October 2012, Pages 54–69
Dynamics of the Laschamp geomagnetic excursion from Black Sea sediments
N.R. Nowaczyk et al.
Investigated sediment cores from the southeastern Black Sea provide a high-resolution record from mid latitudes of the Laschamp geomagnetic polarity excursion. Age constraints are provided by 16 AMS 14C ages, identification of the Campanian Ignimbrite tephra (39.28±0.11 ka), and by detailed tuning of sedimentologic parameters of the Black Sea sediments to the oxygen isotope record from the Greenland NGRIP ice core. According to the derived age model, virtual geomagnetic pole (VGP) positions during the Laschamp excursion persisted in Antarctica for an estimated 440 yr, making the Laschamp excursion a short-lived event with fully reversed polarity directions. The reversed phase, centred at 41.0 ka, is associated with a significant field intensity recovery to 20% of the preceding strong field maximum at ∼50 ka. Recorded field reversals of the Laschamp excursion, lasting only an estimated ∼250 yr, are characterized by low relative paleointensities (5% relative to 50 ka). The central, fully reversed phase of the Laschamp excursion is bracketed by VGP excursions to the Sargasso Sea (∼41.9 ka) and to the Labrador Sea (∼39.6 ka). Paleomagnetic results from the Black Sea are in excellent agreement with VGP data from the French type locality which facilitates the chronological ordering of the non-superposed lavas that crop out at Laschamp–Olby. In addition, VGPs between 34 and 35 ka reach low northerly to equatorial latitudes during a clockwise loop, inferred to be the Mono lake excursion.
Link