Application to K12b components
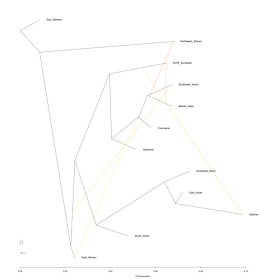
I have applied this idea to the K12b components. Here is the tree with no migration edges, using the Sub_Saharan component as an outgroup:
West Eurasian components that I've labeled "the Six" group together, but the Northwest African one is intermediate between the others and the two African components.
Now, let's allow one migration edge:
Now, the Northwest African component seems derived from what could be called "Indigenous Northwest Africans" but there is a migration edge going to it from a southern Caucasoid population.
Let's allow two migration edges:
Now, there appears to be some gene flow from what appears to be an early Proto-Eurasian population into Southwest Asians. This may be consistent with my idea that the Southwest Asian components represents an amalgam of Neolithic migrants from the "core area" with pre-existing inhabitants of the southern Near East.
With three migration edges:
There now appears to be some gene flow from East Africa to southern Caucasoids. One might speculate that this has something to do with the dispersal of Y-haplogroup E1b1b and/or mtDNA haplogroup M1?
With four migration edges:
There now appears gene flow from the Atlantic_Med into the North_European component. Does that indicate the absorption by the ancestors of the North Europeans of an Oetzi-like substratum?
With five migration edges:
There now appears some input into the Siberian component by a Proto-North European one. This may be related to steppe-related dispersals of northern Caucasoids in Siberia during the Eneolithic and later times, and/or Proto-Europoids like Kostenki and its eastern relatives?
Notice also, that it appears that the North_European input into Siberian precedes the Atlantic_Med input into North_European. So, this is consistent with an eastern origin of North_European which absorbed Atlantic_Med/Oetzi-like populations in Europe and contributed to the East Eurasian native population in Siberia.
Strength of the Edges
The strength of the edges (for the -m 5 run) is:
- 73% of Southwest_Asian -> Northwest_African
- 7% of East_African -> Southwest Asian/Caucasus/Atlantic_Med group
- 42% of Atlantic_Med -> North_European
- 18% Proto-Eurasian -> Southwest Asian
- 19% of North_European -> Siberian
Notice that these are inferred contributions between ADMIXTURE components. Extant populations are composed of different proportions of these components.
Technical Note
Here is how to convert ADMIXTURE output into TreeMix allele counts. This relies on the P and Q files output by ADMIXTURE software.
The P file is an MxK matrix, where:
M: number of SNPS
K: number of components
Suppose one entry in this array is, say, 0.6.
This is consistent with a 6,4 entry in the corresponding TreeMix input file, because the first allele has a frequency of 6/(6+4) = 0.6, and the other allele has a frequency of 0.4 = 4/(4+6).
But, it's also consistent with e.g., 12,8 or 24,16, etc.
You can figure out how many alleles in total to use, by exploiting the Q file, which is an IxK matrix, where
I: number of individuals
K: number of components
If you sum up one of the columns in this array, you get a number of equivalent "individuals" that each ADMIXTURE component corresponds to, based on the original ADMIXTURE run.
The entries in the TreeMix input file are then like this, for the k-th population and m-th SNP
2 * sum(Q[,k]) * P[m, k] , 2 * sum(Q[,k]) * (1-P[m, k])
You might want to round up these numbers, as they may not be exactly integer.
UPDATE (March 13): I've made a script to convert ADMIXTURE output into TreeMix input.






Very interesting.
ReplyDeleteAlthough I would, once again, disagree with the suggestion that the "Proto-Eurasian" gene flow into Southwest Asian is from an aboriginal Arabian population. There is a lack of genetic evidence to support the idea that Arabians descend from an OoA population with a high African affinity to a greater degree than other Eurasians.
East Africans do, however, show signs of proto-Eurasian ancestry (mtDNA L3). I believe this inferred gene flow is most probably caused by East African gene flow, from an East African population that was more similar to Eurasians than your East African cluster. Most likely from the Horn of Africa, or thereabouts. Some earlier experiments by yourself and others have shown that Yemeni Jews, an isolated Arabian population, derive part of their ancestry from a population more distinctly "East African" than East Africans themselves. This is consistent with the finding of deeply rooted East African mtDNA lineages, such as L3x1, in the Yemeni Jewish population. This gene flow probably preceded the spread of some L(xL3) lineages in the Horn of Africa.
A greater amount of samples from East Africa should elucidate this relationship further.
Gene flow is from a population that seems to be ancestral to all Eurasians, hence Proto-Eurasians. Whether that population lived in Africa or not is a different question.
ReplyDeleteActually, this population needs not to be ancestral to Proto-Eurasians; it simply appears to be symmetrically related to all of them.
If I had to wager, I'd say we are dealing with Gulf Oasis denizens that intermixed with incoming Neolithic peoples from the north. These Gulf Oasis denizens are, in turn, the descendants of early Homo sapiens in Arabia and environs, i.e., the people that stayed behind during the Out-of-Arabia event.
You could be right. Only problem is the lack of Y-DNA and mtDNA to support the survival of such a population. Also, I see no particular reason why Arabians that "stayed behind" until the Neolithic would have such a high African affinity.
ReplyDeleteOceanians would also be interesting to include in this kind of experiment, too bad they were not in the K12b. Tishkoff et al. (2009) found that the "African" affinity of Oceanians in ADMIXTURE/STRUCTURE at low K values is related to East Africans.
"So, this is consistent with an eastern origin of North_European"
ReplyDeleteThis can be, but must not.
If I remeber that those recent Y-DNA checks on several thousand years old Ukrainians and even Hungarians (coming back as East Asian) suggested that the border between Europe and Sibiria could have been the eastern border of Poland or something like that.
In such a scenario, would a central European origin of "Northern European" still lead to Sibirians and proto-Northern Europe beeing direct neighbours.
Also, with a more central origin, the spread of quiet large amounts of "Proto NOrthern European" from the Atlantic to the Ural is easier to explain than one massive sweep from the Russian plains to the Atlantic coast.
Another thing that points to an "Explosion" into several directions, from the center is, where those points of maximum STR diversity for R1 clades in Europe are.
Virtually all western European R1b clades have maximum STR diversity in southern Germany or even Czechia (the Italian/French one for example). European R1a apears to have maximum diversity somewhere between southern Poland and Northern Yoguslavia. Wich is virtualy right beside each others and pretty central.
Now imagining, one half goes eastward from the center and pushes the border of Sibiria to the Ural and one goes west from the center.
Teh assumtion that Proto-Northern European bases on the fact that the 2 major clades of R1 are the only haplogroup that has a spread far enough to cover all the space.
I'm quite impressed at the information provided by the algorithm, admittedly. All or almost all the results at m=5 seem consistent with what I could have expected (more or less).
ReplyDeleteThe main doubt I have is how to interpret the weights, notably does "73% of Southwest_Asian -> Northwest_African" mean that 73% of the NW African gene pool is from SW Asia? If so, why does the algorithm not hang North Africans from the WEA macro-population and declare instead a 27% of African admixture?
Otherwise:
1.- the "Proto-Eurasian" admixture with Arabs and such (SW Asian) is consistent with a remnant of the "coastal" OoA via Arabia remixing with backmigrants from South Asia (the "Gedrosia" link) back-flowing to West Eurasia c. 55 Ka ago. They would only mix significantly with SW Asia branch, not taking part in the competition with Neanderthals for the rest of the region. 18% is a nice remnant, roughly consistent with what we can see in mtDNA . Similarly there's a roughly 27% of L(xM,N) in North Africa.
2.- The 7% East African admixture probably correlates with the penetration of Y-DNA E1b.
A major doubt I have is whether the populations mentioned here are real pooled samples or rather, as you seem to love to do, "zombies": artificial synthetic populations mad out of selected components.
PS - The email subscription button seems to not work yet. I believe it's caused by the latest changes by Blogger (incl. the new hateful captcha). In my blogs I have been forced to use the window at bottom format to allow again for an email subscription button to show up.
"Notice also, that it appears that the North_European input into Siberian precedes the Atlantic_Med input into North_European. So, this is consistent with an eastern origin of North_European which absorbed Atlantic_Med/Oetzi-like populations in Europe and contributed to the East Eurasian native population in Siberia."
ReplyDeleteIMO this is consistent with the North_European/Siberian admixture preceding the Northern-European/Atlantic Med admixture in time. The first mix takes place lower down the Northern-European branch, the population moves along the branch with time. I agree this is the steppe dwellers (Gravettian, 22-32k ago). The admixture with the Atlantic med happened later to a population that had moved further away from the root.
The indigenous North African component is interesting. I assume that the strength of the migration is reflected in how easily it manifests?. This would then be the biggest mix. So a very big movement from South West Asia into an indigenous North African population. I suppose that is possible.
(put this in the wrong blogger box initially)
The Beringia land bridge existed 40,000—16,500 years ago.
ReplyDeleteThe Americas seem to have been colonized towards the end of this era (maybe).
Does that make Pickrell and Pritchard 's weird Russian-Maya link evidence that the Maya are descended from a Gravettian-derived people (like the Clovis folk), possibly stuck in the Americas when the land bridge flooded?
This would make the other populations comparatively non-Gravettian (non Clovis?), which is even more interesting.
I wonder what the tree would look like if you eliminated the africans? Presumably this would allow the visualization of more migration events?
There now appears gene flow from the Atlantic_Med into the North_European component. Does that indicate the absorption by the ancestors of the North Europeans of an Oetzi-like substratum? [...] 42% of Atlantic_Med -> North_European
ReplyDeleteD, if the NE component is 42% proto Atlantic Med then why is the NE component still closer to the West Asian component than to AM?
http://2.bp.blogspot.com/-gtp9YiXJjkc/TyfhuRvn8uI/AAAAAAAAEdU/-fgt57DJRio/s1600/thesix_global.png
Is it because the NE component received input from proto AM before AM received input from a pre-existent west Med substratum, the absorption of which later far shifted the AM? The input to the NE component would then not be like Otzi but like the proto AM component before the proto AM absorbed the pre-existing west Med substratum to form the AM Otzi-like component.
Given the closeness of NE to WA away from the Caucasus centre, I would guess that the NE received input from the same pre-existing substratum as did the WA component. (Or could further migrational edges show an input of WA to NE?) The closeness of the NE component to WA would also seem to suggest that the proto-NE component did not absorb much substratum from the pre-existent northen European population (possibly related to the near extinction of Hg. I1?)
The scenario might be that proto NE absorbed some of the proto AM and mixed with the same pre-existing substratum as WA presumably all in west Asia (or with the WA component itself?) and then headed north but did not mix as much with the pre-existing north European substratum. The proto AM (possibly already somewhat mixed with the proto WA component or with the same pre-existing substratum as WA?) meanwhile headed west and mixed heavily with the pre-existing west Med substratum.
Also, I must ask, if the Caucasian components are shifted away from the central Caucasus_component because they have absorbed pre-existing substrata (none of which show up directly in this componential admixture analysis) then can those substrata not be revealed through some componential analysis?
On thinking about it, I am surprised by the separation between the Atlantic Med and Northern Europeans. I would have expected them to have diverged from the same early source population (that progressed on to Caucasus and South West Asia).
ReplyDeleteThis tree seems to be saying that the Atlantic Med flow is comparatively recent development, when I would have expected it to be the oldest to have split off from the group (the coastal movement around the Med to the Atlantic). Could this component be distorted with excessive Caucasian and South West Asian input? Perhaps an admixture of the true early Atlantic Med and a later Caucasoid flow?
The position of the Atlantic Med just looks wrong, it looks like it originated somewhere East of the Caucasus in relatively recent times. This does not seem to fit anything we (think we) know.
It also does not match Pickrell and Pritchard's tree well, which (albeit flawed in its own way) has the Caucasian Adygei associated more closely with the Northern European Russians, and the Europeans (AM and NE) diverging off together after Gedrosians (eg Balochi).
Fascinating, great new tool.
ReplyDeleteAre you going to do some runs focusing on Europe - real or zombie populations (high K, preferably)?
Of course, as always, my own interpretation is more pre-neolithic for Europe (and still consistent with the results), but we will know for sure at a later time.
Just an idea...
ReplyDeleteWhy don't you try to use the Treemix with Chromopainter pops.
Something I forgot to mention is that a most fascinating result of this exercise is that there is NO NEOLITHIC ADMIXTURE apparent anywhere: West Eurasians are a single block hanging from the pan-Eurasian trunk and not a single flow that can be considered Neolithic admixture is apparent.
ReplyDeleteWithin that West Eurasian bloc, the only admixture detected at m=5 is "42% of Atlantic_Med -> North_European", which is rather consistent with Magdalenian expansion or something like that. I know Dienekes claims that all West Eurasian components are Neolithic to some extent but should not this exercise detect some such migrational flows. Should not the pre-Neolithic layer be apparent somehow, the same that a pre-WEA layer is apparent (in various forms) in SW Asia or NW Africa. Isn't there mtDNA U5+U4 in Europe enough to compare with the mtDNA L(xM,N) in North Africa or Arabia?
I think, Dienekes that you may have stumbled here with the evidence of low to zero Neolithic migration in Europe. Or can you detect it at all with some other analysis strategy? It does not look like (and this is a scientific challenge indeed).
Yeah. To me this tree also suggests AM to be the last wave and NE to be the first wave.
ReplyDeleteThats what I read from it:
1. All Caucasoid populations base on one source, wich is in the middle east. (Except for beeing from Africa in the first place)
2. Proto-Northern Europeans beeing the first "Caucasoids" who leave the middle east towards an unknown place. (Either for Central Europe or the Russian/Asian planes is questionable)
Their final destination in Europe beeing Northern Europe + Western Europe (including Northern Italy)
3. The Caucasoids of later Southern India leave the middle east. Roughly at the same time as the Northern Europeans meet Sibirians.
3. Additional DNA flow from Africa leads to seperation of the remaining middle easterners into Northern (Caucasus) and Southern (Southwest Asian). I expect the southern ones to be the ones who are changed by the African input.
The final European destination is Southern Europe. Specially Balkan peninsular and Italy.
4. Those southern middle easterners leave the middle east and become Atlanto-Med. Destination Southern Europe and Western Europe.
------
Something else I think about when comparing AE and NE:
Pigmentation of hair and eyes. In this, NE and AM are the exact oposite sites. The populations with the highest levels of NE possess the highest amounts of alleles that lead to low pigmented hair and eyes (in fact a map of the frequency of the most important "Blue Eye" allele looks identical to maps of "Northern European" admixture components. Almost as if Admixture judges this component just by the existance of this allele.)
The top allele of those was estaminated as 8K-10K years old and originating "Northwest of the Blacksea" by some Danish scientists.
Sardinians are the one Europeans who possess the lowest frequencies of these low pigmentation alleles. And they are the center of AM. Also Oetzi does not possess it aswell.
I find this an interesting additional factor.
Something I forgot to mention is that a most fascinating result of this exercise is that there is NO NEOLITHIC ADMIXTURE apparent anywhere: West Eurasians are a single block hanging from the pan-Eurasian trunk and not a single flow that can be considered Neolithic admixture is apparent.
ReplyDeleteThe entire West Eurasian block is Neolithic or close to it. You can check this easily by looking at the drift parameter. Actually, you don't even have to check this, since the original Fst's of K12b between the components have Neolithic age estimates.
Your insistence on the failed Paleolithic continuity model is admirable, but there is nothing in these results consistent with deep Paleolithic continuity of Europeans.
I think, Dienekes that you may have stumbled here with the evidence of low to zero Neolithic migration in Europe.
Dream on.
2. Proto-Northern Europeans beeing the first "Caucasoids" who leave the middle east towards an unknown place. (Either for Central Europe or the Russian/Asian planes is questionable)
That's one possible interpretation. A different one is that North Europeans have Mesolithic European admixture (consistent with high mtDNA U in eastern Europe, where the North European component is modal), and this inflates their divergence with other Caucasoids.
The drift parameter cannot be used as a measure of absolute age, just relative age. All the tree lines are on different scales, as illustrated by the fact that the migration edges are not perpendicular.
ReplyDeleteThe Fst calculations are heavily biased by the choice of parameters (eg population sizes). They need to be calibrated with real events and facts. Most of the required facts are unknown, and can only be guestimated. I don't place a lot of weight on them. Interesting to play, with but weak and easily distorted.
Well, all this is novel for me and I have not toyed with the program so far, so I did miss the drift parameter till now. Thanks for mentioning it.
ReplyDeleteBut then, based on those drift measures, which markedly make the North European component apart, while the Atlantic-Med is instead hyper-close to the SW Asian zombie, that the North European component is Paleolithic and the Atlantic-Med is Neolithic? That would make some good sense with this graph, better sense than you suggest.
"since the original Fst's of K12b between the components have Neolithic age estimates".
They do not actually: even following your methodology most fall to doubly old ages than Neolithic.
"Your insistence on the failed Paleolithic continuity model is admirable"...
It would not be "admirable" if I'm wrong but I fail to see any clear evidence of the Neolithic model making any sense at all. If I insist is because those defending the Neolithic model fail to make any sense.
The drift parameter cannot be used as a measure of absolute age, just relative age. All the tree lines are on different scales, as illustrated by the fact that the migration edges are not perpendicular.
ReplyDeleteThey can be transformed to absolute age with a standard methodology. You are, of course, free to criticize the methodology _with facts and arguments and numbers_ but not with "it can't be done" pronouncements.
The Fst calculations are heavily biased by the choice of parameters (eg population sizes). They need to be calibrated with real events and facts. Most of the required facts are unknown, and can only be guestimated. I don't place a lot of weight on them. Interesting to play, with but weak and easily distorted.
My own age estimates are reasonable; for example the divergence of the various Eurasian components coincides with the known appearance of AMH throughout Eurasia c. 50-40ka. The divergence of the West Eurasian components coincide with the Neolithic. And, the divergence of the African with the Eurasian components also coincides with Out-of-Africa (or Out-of-Arabia) scenaria about the peopling of the world.
But then, based on those drift measures, which markedly make the North European component apart, while the Atlantic-Med is instead hyper-close to the SW Asian zombie, that the North European component is Paleolithic and the Atlantic-Med is Neolithic? That would make some good sense with this graph, better sense than you suggest.
No, the North European component has the earliest split from the others, and this may correspond to a late Paleolithic/Mesolithic divergence or to a later Neolithic/post-Neolithic divergence coupled with absorption of a Paleolithic substratum.
They do not actually: even following your methodology most fall to doubly old ages than Neolithic.
The ones that have high divergence times are easily explained by admixture, which is evident in the graph, and entirely consistent with my comments. The other dates are all about ~10ka postdating the onset of the Neolithic when populations began diverging as they started expanding from the "core area"
It would not be "admirable" if I'm wrong but I fail to see any clear evidence of the Neolithic model making any sense at all. If I insist is because those defending the Neolithic model fail to make any sense.
We make plenty of sense:
- European ancient mtDNA is entirely unlike modern European mtDNA, being dominated by haplogroup U
- Neolithic Y-chromosomes down to Oetzi's times are lacking the entire R1 clade that makes the bulk of extant European Y-chromosomes
- Oetzi from the Alps is like modern Sardinians and not like modern North Italians or Central Europeans
- Age estimates based on autosomal data coincide with the onset of the Neolithic and are irreconcilable with deep Paleolithic divergence
All these elements point to the idea that there has been an upheaval in the gene pool of Europe over the last few thousands of years. You can cling onto Paleolithic continuity, but there's much more evidence and "sense" in the theory of recent replacement than in your own theories.
At some point wouldn't we expect a migration path from "Gedrosia" to South Asian? Why is this not present?
ReplyDeleteDienekes, shouldn't we see a migration path from "Gedrosia" to "South Asian"? Is there a reason for this absence?
ReplyDeleteThe divergence of the West Eurasian components coincide with the Neolithic.
ReplyDeleteThinking tentatively about this, I guess the only other way to explain this would be to say, "Well, all that means is that West Eurasians were more or less panmictic (unstructued or very loosely structured) until the Neolithic and weren't like that afterwards" (presumably under this paradigm this is an effect of differential transitions to agriculture and sedentism and regional transformations in population sizes).
But of course, the mtdna and craniometry are what (AFIAK from this blog) are what actually show the existence of a population which was replaced and the absence of a panmictic West Eurasian population. And this makes this panmictic explanation unviable and Neolithic replacement the better interpretation of the divergence of the West Eurasian populations, rather than a worse one than breakdown of panmixia (because of course, a pure breakdown of panmixia explanation would not require an "extra" population invisible to the component analysis - at least until we get the Cheddar Man genome [or a similar genome]).
(Of course, a panmictic paradigm wouldn't mean "Our ancestors were here all along since the Paleolithic" but "Our ancestors were here and so were theirs and ours were there as well". But is still a kind of Paleolithic/Mesolithic continuity.)
"They can be transformed to absolute age with a standard methodology. You are, of course, free to criticize the methodology _with facts and arguments and numbers_ but not with "it can't be done" pronouncements."
ReplyDeleteOK then. Let us take the NE-Siberian migration event which I think we agree took place in the Gravettian (22-32k ago). According to your data the NE-Siberian migration event occurred at a drift value of 0.06 for the NE and 0.10 for the Siberians. This is a very big difference for a simultaneous event.
Also we know from remains that the haplogroups like H and V were already well established in North Africa 10,000 years ago. So the NWA-SWA migration event is pre neolithic. So the Atlantic Med (AM) must have split off from SWA significantly earlier than 10,000ky ago (as not that close to the join). And the split off of the other "West Eurasian component", the Northern Europeans, is clearly substantially earlier. These are not neolithic events.
Any signature of gross population movement in the neolithic should show as a migration event from the SWA or the Caucasus into the Atlantic Med and NE (both of whom had already left the cradle of the neolithic). There isn't one at high drifts.
Your Atlantic Med still does not feel right. I am shocked it appears to have broken away so long after Northern Europeans. I expected it to be older. I wonder what this tree would look like if Otzi was used as a possible Atlantic med zombie.
Dienekes said:
ReplyDeleteEuropean ancient mtDNA is entirely unlike modern European mtDNA, being dominated by haplogroup U
Would you care to provide sources where one can see that ancient (Mesolithic/Paleolithic I presumed you meant) mtDNA found in places like England, Iberia, France is entirely dominated by Haplogroup U. I mean not just one person like the Cheddar man, but a set of samples. Your characterization of 20 samples from Russia, Poland, Germany and Lithuania as representative of all the Mesolithic European gene pool is mind-blowing. On the other hand your attitude to dismiss the Mesolithic data from Chandler et al(2005) just because it wasn’t published on a Journal, yet it was presented on an Archeological Congress, is sad. I already showed that if we are to use the effective population size of the French Basques, then it is best to use it to determine the divergence time of the French Basque component as it appears in your ADMIXTURE run back in April, 2011. Given that you claim that the Ne of the Adygei is inflated because of them being admixed, let’s just look at the Fst distance of the different components from the West Asian component at K=10, and K=11.
K=10
http://2.bp.blogspot.com/-VO3qZEQYilY/TbDLtgVFvcI/AAAAAAAADiI/kdvqPTQaO-Q/s1600/ADMIXTURE_10.png
French Basque-West Asian: 0.038
Sardinian-West Asian: 0.038
NE European-West Asian:0.035
NW European-West Asian:0.029
Following your assumption that NE Europeans are more divergent because they absorbed more Paleo/Mesolithic genes due to the prevalence of mtDNA Haplogroup U there today, how does it make sense that both the French Basque component and the Sardinian component are farther apart from West Asian.
K=11
http://2.bp.blogspot.com/-Wk9_ST1CKC4/TbFOmpaIlGI/AAAAAAAADiQ/jhw64bHM1G4/s1600/ADMIXTURE_11.png
French Basque-West Asian: 0.038
Sardinian-West Asian: 0.039
NE European-West Asian:0.036
NW European-West Asian:0.028
Again, would you be so kind to explain why such anomalous results are showing the French Basque and Sardinians to be the most divergent components from the West Asian. Perhaps, drift due to isolation, which might explain the Sardinians, but it doesn’t explain why the French Basque component isn’t equally as divergent from the NW European component(0.03 for K=10, 0.029 for K=11), or the Sardinian(0.032 for both K) as they are from the West Asian component. So it would be awesome if you could explain with your Neolithic colonization scenario how this anomaly came to be.
- Neolithic Y-chromosomes down to Oetzi's times are lacking the entire R1 clade that makes the bulk of extant European Y-chromosomes
Uhmm, If the R1 clade was that of the Paleolithic Europeans, who where known not to have intermingled with the incoming farmers, at least in Western Europe, then why should we expect to find R1 in places that are known to have been colonized by agriculturists.
Maju,
ReplyDeleteAlso, don't forget these are zombie populations. So, even in your paleolithic interpretation, there is neolithic and later admixture: except for the very northeast and Basques, most real European populations have at least 5%-10% Caucasian admixture, and closer to 15%-20% in the eastern Mediterranean and parts Eastern parts of the Balkans.
As to the age estimates, I am also still skeptical, since IMO there is sufficient (but indirect) evidence for a pre-Toba population, so one could normalize the Eurasian split(s) at perhaps 80-90kya, instead.
Would you care to provide sources where one can see that ancient (Mesolithic/Paleolithic I presumed you meant) mtDNA found in places like England, Iberia, France is entirely dominated by Haplogroup U. I mean not just one person like the Cheddar man, but a set of samples. Your characterization of 20 samples from Russia, Poland, Germany and Lithuania as representative of all the Mesolithic European gene pool is mind-blowing.
ReplyDeleteIncorrect.
There is an entire boreal zone of early U-dominance from Cheddar man all the way to Siberia
http://dienekes.blogspot.com/2009/09/some-mtdna-links-between-europe-and.html
There was another Luxembourgian case that I don't believe was mentioned in the above. So, you got Cheddar Man, Luxemburg, Central/Northern Europe, Eastern Europe, all the way to Lake Baikal, where they Europeoid component was apparently monopolized by mtDNA-U
It is really the case that Caucasoid people in Europe from the Atlantic to Siberia were U dominated during prehistory. Samples in southern Europe are lacking, but it is (a) difficult to envisage a separate non-U haplogroup dominated gene pool there that apparently did not spill out into the boreal zone. (b) the inhabitants of northern Europe certainly derived from the south after deglaciation.
In short, all evidence points to a U-dominated European gene pool during the Paleolithic.
On the other hand your attitude to dismiss the Mesolithic data from Chandler et al(2005) just because it wasn’t published on a Journal, yet it was presented on an Archeological Congress, is sad.
No, it is not sad. Some presentation in some regional Iberian conference that failed to make it into a journal is not reliable. It's not even cited in recent published ancient DNA work on Iberia, which ought to give you a clue.
Uhmm, If the R1 clade was that of the Paleolithic Europeans, who where known not to have intermingled with the incoming farmers, at least in Western Europe, then why should we expect to find R1 in places that are known to have been colonized by agriculturists.
The Paleolithic continuity theory postulates that Paleolithic populations adopted farming. Finding that the Neolithic populations were not like the Paleolithic ones is a blow to that theory.
You'd have to imagine that the hypothetical Paleolithic R1 folk were hiding away while the Neolithics inundated the continent with their superior food-producing economy, but somehow they eventually (when? why?) decided to come out of their hideouts and outgrow the Neolithics that had thousands of years in the "growing grops to make lots of babies" business.
So it would be awesome if you could explain with your Neolithic colonization scenario how this anomaly came to be.
No big mystery; these components from earlier analyses are not as reliable as the ones produced more recently, both because I have added many, many populations during the last year, and also because I am now systematically accounting for presence of relatives in all source populations.
As to the age estimates, I am also still skeptical, since IMO there is sufficient (but indirect) evidence for a pre-Toba population, so one could normalize the Eurasian split(s) at perhaps 80-90kya, instead.
There are no AMH in Europe or West Asia at 80-90ka so it is ridiculous to claim that divergence between Eurasians began that early.
"There are no AMH in Europe or West Asia at 80-90ka so it is ridiculous to claim that divergence between Eurasians began that early."
ReplyDeleteYou yourself are flirting with the idea of AMHs in SW Asia at roughly around that time. Of course there were none in regions dominated by Neanderthals. But S Asia and SE Asia are a different story. Toba and the ensuing 10,000 years cold/dry period could have well solidified splits of AMH populations that occurred in the preceding tens of thousands of years while S Asia and SE Asia were settled, by preventing efficient admixture - while further migrations separated the populations even more. All in the subcontinent, SE Asia, and surroundings. No need to evoke West Asia or Europe, at that point. Populations separate before they arrive at their destination.
You yourself are flirting with the idea of AMHs in SW Asia at roughly around that time.
ReplyDeleteYes, but these were Ur-Eurasians, prior to their split. The split of these Ur-Eurasians occurred when they left their cradle (whether it was Africa or Arabia), and modern humans start appearing simultaneously around the world around 50-40ka in places like Europe and Australia.
Also Fst's e.g., between ASI and other Eurasian components don't appear to be very large, they're the same order as other intra-Eurasian Fst's. So, if you find a South Asian split pre-Toba by tweaking with the parameters of the model that would also affect all the other dates in Eurasia.
"This tree seems to be saying that the Atlantic Med flow is comparatively recent development, when I would have expected it to be the oldest to have split off from the group (the coastal movement around the Med to the Atlantic)."
ReplyDeleteIf the coastal movement around the atlantic was relatively small in number - not surprising given the distance from the source (assuming the original source was the eastern med or a secondary colony of same somewhere along the med coast) - and they introduced their domestic animals to the locals they might have produced a hybrid forager-pastoralist population which - if it could support a higher pop. density than foraging alone would expand from the west / northwest. So you get the anatolian tortoise slowly coming up the danube from the SE to the NW while the sea peoples hare that went round the atlantic coast sparked a backflow going NW to SE.
With the R1a movement as a third element.
***
Personally i think this is likely to be a recurring theme of the neolithic farmer expansion. The first farmers would have to hop from one place where the terrain and climate were suitable to the next.
In many of the farming settlements the pastoralist element of the package could be extracted separately and used on nearby terrain that wasn't viable for crops at the time.
So think the farmers would have created a layer of pastoralists around each seed spot who in time would become their conquerors - from Sumer / Akkad onwards.
So, if you find a South Asian split pre-Toba by tweaking with the parameters of the model that would also affect all the other dates in Eurasia.
ReplyDeleteYes, of course. ;)
Dienekes said:
ReplyDeleteThere was another Luxembourgian case that I don't believe was mentioned in the above. So, you got Cheddar Man, Luxemburg, Central/Northern Europe, Eastern Europe, all the way to Lake Baikal, where they Europeoid component was apparently monopolized by mtDNA-U
I take it you are choosing to deliberately ignore part of what I wrote. I said if you had any proofs that places like England, France, Iberia were dominated by mt-DNA Haplogroup U in pre-Neolithic times, and I explicitly said that citing Cheddar man, or the Luxembourg case, all single instances is nowhere near conclusive proof that U was dominant in Western Europe. In fact, there is a Paleolithic remain from Iberia which turned out to be U, yet, there is a saying that absence of evidence is not evidence of absence. Of course, there will always be skepticism from those who do not want to accept the reality, and that’s why nothing is 100% certain, and that’s why we rely on statistics. Now the fact that you are trying to portrait a single sample from England as evidence of mt-DNA U dominance in England, or the same thing for Luxembourg is astonishing. Again, I’m not saying that if 20 remains are analyzed in England and they all turned out to be mt-DNA U or mostly mt-DNA U and H was absent, that I wouldn’t accept that most likely England’s pre-Neolithic mt-DNA pool was U dominated, but to make such conclusion from a single sample is just bogus.
In short, all evidence points to a U-dominated European gene pool during the Paleolithic.
In short, the little evidence that we have attest for the presence of mt-DNA Haplogroup U in pre-Neolithic Germany, Poland, Lithuania and Russia, as well as, mt-DNA J and T2e (both considered Neolithic lineages). All evidence thus far recovered isn’t conclusive enough to make the assertion that mt-DNA U was dominant in the European gene pool during the Paleolithic.
No, it is not sad. Some presentation in some regional Iberian conference that failed to make it into a journal is not reliable. It's not even cited in recent published ancient DNA work on Iberia, which ought to give you a clue.
Well has the ever been a paper published on the mt-DNA of the Cheddar Man, or was the paper on the Luxembourg finding published in a mainstream English journal. You make a good case of a special pleading fallacy, but that doesn’t work on me. Why should a paper on ancient DNA from Catalonia talk about Mesolithic DNA from Portugal, in fact, as far as I’ve seen, all recently published aDNA papers labeled mt-DNA H as pre-Neolithic, while all the wild geese(X, N1a, etc) found in Neolithic sites was labeled as Neolithic mt-DNA. That paper was published in a Spanish Journal, last time I checked it was a conference of the Neolithic in Iberia, and you will gladly take any papers published in the AJPA conference if they were available, so the double standard is indeed sad. The fact, that whenever the findings go against your pre-conceived notions, they are labeled as unreliable is sad, yet the same standards are not applied when those findings support your pre-conceived notions.
Continued...
ReplyDeleteThe Paleolithic continuity theory postulates that Paleolithic populations adopted farming. Finding that the Neolithic populations were not like the Paleolithic ones is a blow to that theory.
You'd have to imagine that the hypothetical Paleolithic R1 folk were hiding away while the Neolithics inundated the continent with their superior food-producing economy, but somehow they eventually (when? why?) decided to come out of their hideouts and outgrow the Neolithics that had thousands of years in the "growing grops to make lots of babies" business..
When did I say that Paleolithic populations adopted farming, in fact, the one thing I said is why should we expect to find y-DNA Haplogroup R1b-M269 in the Germany during the Neolithic, if it is known that the region was settled by agriculturists form elsewhere, or the same thing applies to Catalonia, and Treilles, France. You do point out to a very good question: When and how did R1b come to be the majority haplogroup in Western European areas that were dominated by Haplogroup G? I agree that replacement took place; however, that says nothing about the status of R1b in Europe. So according to you, only folks that came from the East were capable of creating new technology. The pre-Neolithic Europeans were too dumb to learn anything, yet they managed to survive an Ice Age. All I’m saying is that until further proof, all we can do is speculate about the situation, and that yeah, there was population replacement, now the scale of it, is unknown. The geographic distribution of where this replacement hit the hardest and whether expansions of pre-Neolithic European taking refuge in other areas affected that Neolithic expansion is a very different story, which with any luck we will one day come to understand. In the meantime I prefer to look at the data and come up with my own conclusions.
No big mystery; these components from earlier analyses are not as reliable as the ones produced more recently, both because I have added many, many populations during the last year, and also because I am now systematically accounting for presence of relatives in all source populations.
Of course Dienekes, if you would go as far as applied Ad Hominems to your own experiments if they do not produce the desired results, then there is no point in arguing. What exactly determines the reliability of a component, perhaps you could give us actual numbers of the residual error during that run, and if it orders of magnitude higher than the latest experiments then I’ll take it as a reasonable argument. Now your opinion of what constitutes a “real” component and what doesn’t, isn’t reliable; moreover the populations in questions are populations from HGDP so there is no reason to try to apply the presence of relatives in those populations when you are still using them today as you did back then.
I take it you are choosing to deliberately ignore part of what I wrote. I said if you had any proofs that places like England, France, Iberia were dominated by mt-DNA Haplogroup U in pre-Neolithic times, and I explicitly said that citing Cheddar man, or the Luxembourg case, all single instances is nowhere near conclusive proof that U was dominant in Western Europe.
ReplyDeleteThere are no such things as "proofs" because one can never disprove the presence of a particular haplogroup (one would have to sample the entire population to achieve that, which is impossible).
U occurs at frequencies of, say 10-20% at most in most of Europe. So, if you draw two samples from Western Europe and they both turn out to be U, then the probability of that happening, if the Paleolithic frequency was, say 20%, is 0.2*0.2 = 0.04.
Given that Western Europe is not isolated from the rest of Europe, and we have evidence for total U dominance in Central/Eastern Europe among pre-agriculturalists, there is very strong probability that what I am saying is correct, and very strong probability that "Paleolithic continuity" is wrong.
Well has the ever been a paper published on the mt-DNA of the Cheddar Man, or was the paper on the Luxembourg finding published in a mainstream English journal.
I don't care if it's an "English" journal or not. Also, a priori one is inclined to accept facts that harmonize with the rest of the known facts and to doubt facts that do not. The Cheddar/Luxembourg facts are consistent with what we know about European DNA. They are also a priori more trustworthy because they are from a colder region.
In any case, the road is open for anyone who wants to sample Paleolithic Europeans to follow stringent quality protocols and to advance evidence for whatever theory they want. The evidence as it stands argues for substantial changes across Europe in prehistory.
So according to you, only folks that came from the East were capable of creating new technology. The pre-Neolithic Europeans were too dumb to learn anything, yet they managed to survive an Ice Age.
ReplyDeleteI don't know how smart or dumb they were, and that is irrelevant. They had better tech, it doesn't matter one iota how they got it. Farmers outcompete foragers everywhere in the world, except in the fringes where their technological package of crops and animals is unsuitable. People with iron and bronze weapons outcompete people with stone weapons. People with increased social complexity outcompete thinly distributed bands of foragers. It's just the way the world works.
Of course Dienekes, if you would go as far as applied Ad Hominems to your own experiments if they do not produce the desired results, then there is no point in arguing.
You are using "ad hominem" incorrectly.
The later results are more reliable because they are based on larger datasets and a uniform method of pruning relatives as I have said. Anyone who has worked with ADMIXTURE knows that population-specific components emerge at high enough K, and these components do not necessarily have much to do with actual prehistory. There is an Ashkenazi Jewish component that emerges at high K, but we don't have to believe that Ashkenazi Jews have been isolated from the rest of mankind since Paleolithic times.
I agree that replacement took place; however, that says nothing about the status of R1b in Europe.
R1b in Europe is closely related to R1b in Asia. It cannot have become separated from Asian R1b in a Paleolithic time frame. Even using the "evolutionary mutation rate" one gets dates in the last 10,000 years for Asian/European R1b divergence. Moreover, it's lacking in European ancient DNA samples down to quite recent millennia. We now have many Neolithic-to-Copper Age samples from Europe and no R1b. Once again, we can't exclude the presence of R1b in that timeframe, but it is very unlikely, and increasingly unlikely with each new sample. We can certainly already claim that, if it existed there at all, it was present at a much lower frequency.
There are no such things as "proofs" because one can never disprove the presence of a particular haplogroup (one would have to sample the entire population to achieve that, which is impossible).
ReplyDeleteU occurs at frequencies of, say 10-20% at most in most of Europe. So, if you draw two samples from Western Europe and they both turn out to be U, then the probability of that happening, if the Paleolithic frequency was, say 20%, is 0.2*0.2 = 0.04.
I see we are now engaging in Reductio Ad Absurdum. By ignoring certain parts of my post you tried to engage me in a fallacious way, perhaps you missed it when I said:
“…Again, I’m not saying that if 20 remains are analyzed in England and they all turned out to be mt-DNA U or mostly mt-DNA U and H was absent, that I wouldn’t accept that most likely England’s pre-Neolithic mt-DNA pool was U dominated, but to make such conclusion from a single sample is just bogus.”
You know as well as I do that one cannot make any feasible conclusions off a single sample. So assuming that the mt-DNA U occurs at frequency of 10-20% in both Luxembourg or Cheddar Cave region in the UK, and assuming those frequencies haven’t changed since the Paleolithic times (Very unrealistic), the probability of getting a single instance of U in both places is independent, therefore you can’t really multiply them for once. Secondly, any conclusions made from one sample are simply unrealistic, as they would be no standard deviation, no variance, nothing.
I don't care if it's an "English" journal or not. Also, a priori one is inclined to accept facts that harmonize with the rest of the known facts and to doubt facts that do not. The Cheddar/Luxembourg facts are consistent with what we know about European DNA. They are also a priori more trustworthy because they are from a colder region.
A priori doesn’t apply in this case because there isn’t any statistically significant sample from England/Luxembourg, and by using the 20 samples from Germany, Poland, Lithuania, and Russia what you are doing is a hasty generalization, and trying to invoke appeal to tradition, although I don’t see much tradition is this case.
The evidence as it stands argues for substantial changes across Europe in prehistory.
No it doesn’t, the evidence as it stands isn’t conclusive enough to argument substantial changes across Europe in prehistory. At best you can argue that the modern day people living in Catalonia, Spain, Treilles, France have suffered a population replacement at least paternally in the last 5000 years.
Moreover, it's lacking in European ancient DNA samples down to quite recent millennia. We now have many Neolithic-to-Copper Age samples from Europe and no R1b. Once again, we can't exclude the presence of R1b in that timeframe, but it is very unlikely, and increasingly unlikely with each new sample. We can certainly already claim that, if it existed there at all, it was present at a much lower frequency
I find it interesting your characterization of three ancient samples as many. So do you think that if we sample at random 3 individuals from Germany, 7 from Catalonia, and 20 from Southern France we ought to get the y-DNA genetic profile, that if we sampled 200 from each place? Moreover your last statement is a great example of a Gambler’s fallacy.
the probability of getting a single instance of U in both places is independent, therefore you can’t really multiply them for once.
ReplyDeleteYou can multiply them because they are independent. It's not rocket science. The probability that you get a 3,3 in two independent throws of a dice is 1/6 * 1/6 = 1/36.
I find it interesting your characterization of three ancient samples as many.
We have Oetzi, 3 from LBK, several from Iberia and several from France.
Suppose you only had exactly 3 individuals (which is worse than what u actually have). Then, if there was, say, 50% R1b in Western Europe at the time, you have 1/8 probability of getting none in three tries.
The odds are in my favor.
You can multiply them because they are independent. It's not rocket science. The probability that you get a 3,3 in two independent throws of a dice is 1/6 * 1/6 = 1/36.
ReplyDeleteFair enough!!! My mistake!!! Again the problem is that we can’t really make any conclusions to what the probabilities of finding mt-DNA U in a single sample from ancient England or Luxembourg is.
As for the Y-DNA Haplogroups, again why should we expect to find R1b in places that were colonized, if the conclusion you are trying to arrive is that the frequency and distribution of R1b 5000 ybp in Europe is not the same as today, I agree. However, until we test more places we can say that its frequency was relatively low. Up until recently the frequency of lactose tolerance allele T was nonexistent in all the Neolithic samples tested from France, Catalonia, and Germany. If we apply the same approach to it, as you are trying to apply to R1b then the sample found in SJAPL would have been also zero T, yet it was ~27% if a recall correctly, once more why we cannot make hasty generalizations.
PS: The odds aren’t in anyone favor, we’ll have to wait and see my friend.
Again the problem is that we can’t really make any conclusions to what the probabilities of finding mt-DNA U in a single sample from ancient England or Luxembourg is.
ReplyDeleteThe frequency in ancient England is an unknown that can be estimated based on the knowns.
High frequency (what I call U-dominant population) is much more probable than present-day-like frequency.
. If we apply the same approach to it, as you are trying to apply to R1b then the sample found in SJAPL would have been also zero T, yet it was ~27% if a recall correctly, once more why we cannot make hasty generalizations.
Again, read my above comments. You can never say that the frequency is zero, because you can't sample all individuals. Prior to the recent discovery of lactase persistence, we would have guessed that we'd find it in a much lower frequency than today, and ~27% or whatever it was _is_ much lower. We certainly don't expect to never find it; we are bound to find it eventually, and when/where we do we will get a data point binding its time of arrival.
This comment has been removed by the author.
ReplyDeleteVery good timing.
ReplyDeletehttp://dienekes.blogspot.com/2012/03/neolithic-expansions-how-european.html
In the past we have just agreed to disagree on this.
No, I explained to you why you are wrong, and you repeat the same crap. Fortunately, we now have a good paper which shows how H-bearing farmers assimilated the U-bearing foragers.
(corrected)
ReplyDeleteIf you look at the actual remains most are more likely to be H than U (CRS).
http://www.buildinghistory.org/distantpast/ancientdna.shtml
U is almost entirely found in the north in burials when the most likely death rituals were air and fire (no remains). They could well be a minority group.
Two of the Paglicci remains are most likely H. The Red "Lady" of Paviland (Wales) is most likely H (CRS).
The oldest undisputed mitochondrial remains from the south are the 10k old H just across the water in North Africa. To early to be neolithic in Europe.
The transitional remains in central Europe tend to be labled paleolithic by if they are rich in U and neolithic if they are not in an exercise of blatant bias.
In the past we have just agreed to disagree on th
"Fortunately, we now have a good paper which shows how H-bearing farmers assimilated the U-bearing foragers."
ReplyDeleteAlways better to actually read the paper thoroughly before proclaiming its virtues. This paper extols an expansion starting 13kya and ending by about 7kya. Before the neolithic arrived in Europe.