- The Atlantic_Med component is frequent in northwestern Europe
- The North_European component is dominant in northeastern Europe and forays into Siberia
- The Caucasus component is dominant in the Caucasus and forays into Central Asia
- The Siberian component is dominant in North Asia and forays into Europe
- The East_Asian component is frequent in East Asia and forays into North Asia
Russian_D, Polish_D, German_D, Finnish_D, Swedish_D, Mixed_Slav_D, Norwegian_D, Lithuanian_D, Japanese_D, Daur, French, French_Basque, Hezhen, Japanese, Oroqen, Russian, Sardinian, Yakut, CEU30, JPT30, Belorussian, Chuvashs, Hungarians, Lithuanians, Romanians, Selkup, Evenk, Tuva, Yukagir, Nganassan, Dolgan, Buryat, Mongol, FIN30, Kent_1KG, Bulgarians_Y, Ukranians_Y, Mordovians_Y
Additionally, a sample of 30 Yoruba from the HapMap-3 was used as an outgroup.
TreeMix analysis
The TreeMix analysis was performed with default parameters, and allowing for a different number of migration edges.
Nomenclature: The direction of gene flow is best seen in the figure and/or associated treeout files.
For the text, I will put in (), the common ancestor of two populations, e.g., (French_Basque,Sardinian) and also as (X, *) the tree rooted at a particular node X, e.g., (Buryat, *)
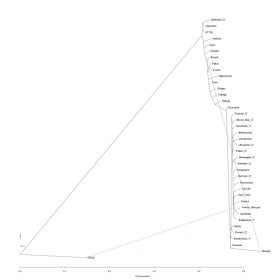
0 migration edges:
The West and East Eurasian clusters are identified, with some populations with likely admixture being placed closer to the Eurasian root.
1 migration edge:
64% from (Sardinians/Basques) to Yoruba; this is difficult to interpret, but there has been evidence in the past that Africans and West Eurasians share more ancestry than Africans and East Asians do. In the linked post, I proposed a major episode of back-migration into Africa, and it is perhaps this that is being captured by this migration edge: Sardinians/Basques are the only two South-West Eurasian populations included, and any back-migration into Africa must have originated in the southern parts of West Eurasia.
Such a high level of back-migration may in fact be plausible, since Yoruba are a predominantly Y-haplogroup E bearing population, and the origin of the DE clade of the human Y-chromosome phylogeny is up in the air with both an African and Eurasian case having been advanced. Personally, I favor the Eurasian case, since within the CT clade, we have two subclades: CF (Eurasian) and DE (Eurasian/African).
Such a high level of back-migration may in fact be plausible, since Yoruba are a predominantly Y-haplogroup E bearing population, and the origin of the DE clade of the human Y-chromosome phylogeny is up in the air with both an African and Eurasian case having been advanced. Personally, I favor the Eurasian case, since within the CT clade, we have two subclades: CF (Eurasian) and DE (Eurasian/African).
Interestingly, John Hawks has recently discovered an unanticipated excess of "Neandertal ancestry" in Yoruba. This may also point to a back-migration into Africa and/or admixture of a group of Africans related to Eurasians (whom I've called Afrasians), with groups of Africans (Palaeoafricans) that split before the H. sapiens/H. neandertalensis common ancestor.
There is, however, another detail in the figure that may have escaped your notice: there is now about 0.5 worth of drift in the figure (left-to-right) as opposed to only 0.12 in the tree without migration edges. So, perhaps what we are seeing is indeed the first sign of admixture between modern and archaic humans in Africa, which has been made more likely by recent anthropological discoveries.
It's not clear to me whether TreeMix has stumbled onto something important or not, but it is certainly worth keeping in mind that the above model fits the data better than the simple tree model. Moreover, TreeMix attempts to reverse the polarity of migration edges, and -apparently- the (Sardinian, French_Basque)-to-Yoruba edge is preferable to the reverse.
So, we should keep our minds open to the possibility that the greater similarity of West Eurasians to Africans is not the result of multiple Out-of-Africa waves, one of which affected only West Eurasians, but of an Into-Africa back-migration from West Eurasia.
So far, tree-based models have focused on how diverse African groups are, and hence, the reduced diversity of Eurasians has been interpreted as an Out-of-Africa bottleneck that carried a subset of African variation into Eurasia.
But, there is an alternative interpretation of the evidence, namely that African groups are diverse because they carry a superset of ancient Into-Africa variation, with the African-specific part of their variation being the result of admixture with pre-existing African hominins. Such a scenario cannot be captured by tree models, but is apparently considered and not rejected by TreeMix which allows for lateral gene flow. Let's wait and see what new things come from full genome sequencing.
So, we should keep our minds open to the possibility that the greater similarity of West Eurasians to Africans is not the result of multiple Out-of-Africa waves, one of which affected only West Eurasians, but of an Into-Africa back-migration from West Eurasia.
So far, tree-based models have focused on how diverse African groups are, and hence, the reduced diversity of Eurasians has been interpreted as an Out-of-Africa bottleneck that carried a subset of African variation into Eurasia.
But, there is an alternative interpretation of the evidence, namely that African groups are diverse because they carry a superset of ancient Into-Africa variation, with the African-specific part of their variation being the result of admixture with pre-existing African hominins. Such a scenario cannot be captured by tree models, but is apparently considered and not rejected by TreeMix which allows for lateral gene flow. Let's wait and see what new things come from full genome sequencing.
2 migration edges:
The (French_Basque/Sardinian)-to-Yoruba edge persists (64%) and a new edge was added from (Buryat, *)-to-Mongol (85%). The "Mongol" sample consists of Siberian Mongols described by Rasmussen et al. (2010). An inspection of their K12b population portrait indicates that they do, in fact, have West Eurasian admixture, which according to the K12b spreadsheet amounts to about 18% in total.
3 migration edges:
The aforementioned (French_Basque/Sardinian)-to-Yoruba (64%) and (Buryat,*)-toMongol (85%) edges persist, and now we have a 68% Nganasan-to-Selkup edge.
These are the two Siberian Uralic populations in the dataset. This seems to parallel the K12b results, as Selkups have a North_European element which the Nganasans (Uralic speakers from the Arctic coast of Central Siberia lack), so we are seeing the hybridity of the Selkups here, who, like the Mongol sample are partly of West Eurasian ancestry.
4 migration edges:
The aforementioned (French_Basque,Sardinian)-to-Yoruba (64%), (Buryat,*)-to-Mongol (84%), and Nganasan-to-Selkup (68%) persist, and now we have a 89% (Buryat, *)-to-Tuva edge. According to the K12b the Tuva have 13.3% West Eurasian admixture, so again we have reasonably good agreement between TreeMix and ADMIXTURE.
Interestingly, the non-"eastern" component of Selkups and Tuvans now forms a clade. It seems that a Nganasan-like and a (Buryat, *)-like population have converged into southern Siberia, absorbed a common local element and became the Selkup and Tuva respectively.
5 migration edges:
The aforementioned (French_Basque,Sardinian)-to-Yoruba (64%), (Buryat,*)-to-Mongol (85%), Nganasan-to-Selkup (68%) persist, and 90% (Buryat, *)-to-Tuva persist, and now we have a new 18% Oroqen-to-(Yakut, Evenk) edge. The Oroqen and the Evenk are Tungusic speakers, whereas the Yakut are Turkic people from northeastern Siberia, having migrated there from the vicinity of Lake Baikal during the last millennium.
6 migration edges:
6 migration edges:
The aforementioned (French_Basque,Sardinian)-to-Yoruba (64%), (Buryat,*)-to-Mongol (85%), Nganasan-to-Selkup (68%), 90% (Buryat, *)-to-Tuva persist, 18% Oroqen-to-(Yakut, Evenk), persist, and a new 16% Nganasan-to-Oroqen edge appears. Interestingly, this has allowed the Oroqen and Hezhen to now form their own clade, which makes sense as these are both Tungusic speakers from northeastern China. The other Tungusic population, the Evenk group with the Turkic Yakut: what they share in common is that they both share origins close to Lake Baikal in Siberia.
7 migration edges:
The aforementioned (French_Basque,Sardinian)-to-Yoruba (64%), (Buryat,*)-to-Mongol (85%), Nganasan-to-Selkup (68%), 90% (Buryat, *)-to-Tuva persist, 18% Oroqen-to-(Yakut, Evenk), 16% Nganasan-to-Oroqen edges persist, and there is a new 81% Evenk-to-Yukagir edge. The remainder of the Yukagirs' ancestry is derived from the West Eurasian tree. The Yukagir language is rather mysterious, with some links to Uralic having been postulated. Here it pays off to look at the population portraits, since it is apparent that -unlike the Selkup- their West Eurasian ancestry is limited to a few individuals.
It is fairly interesting that Russian anthropologists placed the Yukagirs in the Baikal group of the Central Asian race, the same as the Evenks, who are their biggest donors. So, Yakuts, Evenks, and Yukagirs all seem to share the same Baikal-type of origin.
8 migration edges:
7 migration edges:
The aforementioned (French_Basque,Sardinian)-to-Yoruba (64%), (Buryat,*)-to-Mongol (85%), Nganasan-to-Selkup (68%), 90% (Buryat, *)-to-Tuva persist, 18% Oroqen-to-(Yakut, Evenk), 16% Nganasan-to-Oroqen edges persist, and there is a new 81% Evenk-to-Yukagir edge. The remainder of the Yukagirs' ancestry is derived from the West Eurasian tree. The Yukagir language is rather mysterious, with some links to Uralic having been postulated. Here it pays off to look at the population portraits, since it is apparent that -unlike the Selkup- their West Eurasian ancestry is limited to a few individuals.
It is fairly interesting that Russian anthropologists placed the Yukagirs in the Baikal group of the Central Asian race, the same as the Evenks, who are their biggest donors. So, Yakuts, Evenks, and Yukagirs all seem to share the same Baikal-type of origin.
8 migration edges:
There is now a 64% Sardinian-to-Yoruba edge, a 16% Oroqen-to-Yukagir edge, 20% (Buryat, *)-to-(Yakut, Evenk), and a 24% Nganasan-to-Chuvash edge, 29% Oroqen-to-(Yakut,Evenk) edge, 88% (Buryat, *)-to-Tuva, 62% Nganasan-to-Selkup, 85% (Buryat, *)-to-Mongol.
The tree has been rather re-organized, with two main Siberian groups identified: an eastern group (Hezhen, Daur, Oroqen, Buryat), and a central group (Yukagir, Dolgan, Nganasan, Yakut, Evenk, Selkup). The Chuvash, predominantly Europeoid Turkic speakers from Russia show evidence of gene flow from the central group as well, whereas the Selkup, Uralic speakers from Siberia, who belong to the central group, show evidence of gene flow from Europe.
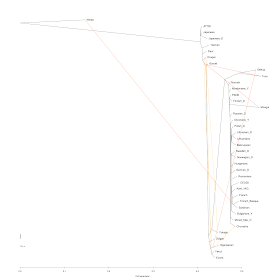
9 migration edges:
64% (French_Basque,Sardinian)-to-Yoruba, 85% (Buryat, *)-to-Mongol, 68% Nganasan-to-Selkup, 92% (Buryat,*)-to-Tuva, 14% Oroqen-to-(Yakut,Evenk), 14% Nganasan-to-Oroqen, 82% Yakut-to-Yukagir, 90% Evenk-to-Dolgan, 13% Hezhen-to-(Nganasan, *).
10 migration edges:
64% (French_Basque, Sardinian)-to-Yoruba, (85% Nganasan, *)-to-Mongol, 68% Nganasan-to-Selkup, 92% (Nganasan,*)-to-Tuva, 15% Oroqen-to-(Yakut,Evenk), 15% Nganasan-to-Oroqen, 82% Yakut-to-Yukagir, 90% Evenk-to-Dolgan, 43% Hezhen-to-Buryat, 14% Sardinian-to-Bulgarian.
I will stop at this point. I may add more migration edges later to this post, but I'm tired of typing this stuff.
You can download all the plots and *.treeout files here.
UPDATE (March 20): I have repeated the experiment with HGDP San, rather than Yoruba as the outrgroup:
There is now a 63% migration edge from (Basque, Sardinian) to San.
UPDATE (March 20): I have repeated the experiment with HGDP San, rather than Yoruba as the outrgroup:
There is now a 63% migration edge from (Basque, Sardinian) to San.











"64% (French_Basque, Sardinian)-to-Yoruba"
ReplyDeleteDoes this mean that, according to this analysis, the Yoruba are 64% European? That's what the Treemix paper seems to say: they'd be only 1-w=1-0.64=0.36=36% "Africans".
This does not make any sense on light of everything else we know and makes me think of totally dumping this kind of analysis. However it may well mean that Treemix is finding too much diversity in the Yoruba and needs of a "migration", maybe representing the ancient L2"6 East African population as distinct from the earliest Central-West African or proto-Pygmy and proto-Fulani L1 populations.
But then why not to draw the origin at the root of the Eurasian subtree?
If my memory is correct, you produced another tree earlier where a Sardinian-originated admixture arrow made up 70% of North African gene pool. This should be related.
Still it generates more confusion than answers. The algorithm seems to need fine tuning or something.
>> Does this mean that, according to this analysis, the Yoruba are 64% European? That's what the Treemix paper seems to say: they'd be only 1-w=1-0.64=0.36=36% "Africans".
ReplyDeleteNot necessarily European, Sardinians and Basques are probably the closest to the Eurasian element that may have contributed to the back-migration into Africa.
>> This does not make any sense on light of everything else we know and makes me think of totally dumping this kind of analysis.
Well, we know that there is "Southern" (K7b) influence in North and East Africa. Henn et al. (2012) postulated substantial back-migration to North Africa and absorption of an indigenous "Maghrebi" component.
If my two-desert theory is correct and H. sapiens originated in the Sahara or thereabouts, it's quite possible that it would be long-diverged from other archaic African hominins.
When the back-migration to Africa took place (bearing ancestral DE chromosomes), the incoming population admixed with indigenous H. sapiens in North Africa, as well as other hominins south of the Sahara. The latter would have been archaic humans like Iwo Eleru and Ishango.
If my memory is correct, you produced another tree earlier where a Sardinian-originated admixture arrow made up 70% of North African gene pool. This should be related.
Yes, I think it is. The back-migration may have affected all of Africa, leading to the disappearance of the archaic forms as they intermixed with the back-migrants. The main difference between North and Sub-Saharan Africa, is that in the North they found Irhoud-like early H. sapiens, and in Nigeria they found Iwo-Eleru-like archaic forms.
This would also be consistent with the finding that Africans have about 4 times more effective population size than non-Africans
ReplyDeletehttp://dienekes.blogspot.com/2011/09/can-we-please-abandon-concept-of.html
Apparent effective size is reduced in a bottleneck (traditional OoA theory) and increased by admixture between divergent populations.
The OoA theory proposes a biblical-level bottleneck OoA, whereas the back-to-Africa theory proposes population structure and admixture between divergent African populations.
I still would like you to do a similar analysis concentrating on Europe...
ReplyDeleteNice analysis,
ReplyDeletebut I am not convinced about the Europe Yoruba migration even if it is not impossible.
Could you gently provide more details about your two deserts theory? It sounds interesting but i cannot fully get it.
Just to know...How long is the analysis (and how may Snps)?
Did you obtain frequencies from plink?
It woul be intresting to see if a similar migration exist also in other W.African populations e.g Bantu.
Thanks again.
Could this be related with the back migrations that carreid R1b in to Sub-Saharan African ? On the other hand, I think the Tree is using the Sardinian as a proxy of the closest thing to North-Africa, due to the lack of any north-african sample here. Add a North-African population, and Im sure the introgression of sardinians will disappear.
ReplyDeleteYES!!!
ReplyDeleteI predicted this 2 years ago!!!
See this post:
http://dienekes.blogspot.com/2010/03/digging-deeper-into-east-african-human.html
pconroy said...
Andrew,
Good questions!
I predict that as more genetic association studies are done, Y-DNA haplogroup E will be revealed to be Eurasian, with only Y-DNA A and B as African in origin.
The interesting question thus, for me, would be which group or groups brought E to Africa and from which direction. I'd further imagine that there could have been 2 sources of agriculture in the Near East, one centered in Southern Anatolia, Northern Syria, the other in the Natufian area, and adjacent areas of North East Egypt.
What do you guys think?
Friday, March 12, 2010 1:52:00 AM
pconroy said...
Ashraf,
AFAIK whenever agriculture spread, it was via male agriculturalists' sons moving to the hunter-gatherer frontier and starting new agricultural communities. So if Eurasian carrying Y-DNA E moved into the Sahara, then the Sahel region, then West Africa proper, the population would initially be Middle Eastern in genotype and phenotype, then with demic expansion and some assimilation of the daughters of hunter-gatherers, become more admixed. Moving to West Africa would favor selection for darker skin and other local adaptations picked up from the native Hunter-Gatherer substrate. So there is a dilution of the Middle Eastern genotype, but maybe a replacement of the Phenotype. and eventually the male agriculturalist on the "wave of advance" are substantially African by descent, but their Y-DNA is still Middle Eastern or more generally Eurasian.
Friday, March 12, 2010 5:47:00 PM
ReplyDeleteInterestingly, John Hawks has recently discovered an unanticipated excess of "Neandertal ancestry" in Yoruba. This may also point to a back-migration into Africa and/or admixture of a group of Africans related to Eurasians (whom I've called Afrasians), with groups of Africans (Palaeoafricans) that split before the H. sapiens/H. neandertalensis common ancestor.
You don't think the Luhya, who are partially East African, would be more likely to have been affected by such a back-migration? And yet, they have a lower similarity with Neanderthals than the Yoruba.
You talk a lot about "North Africans" and "Sub-Saharan Africans" in your post, but fail to give much attention to East Africans. East Africa isn't just the most probable origin of our species, but also the most likely source of the OoA migration (mtDNA L3).
It makes very little sense for African L3(xM,N) to be the result of Eurasian back-migrations when the various African L3 lineages derive from the same root as M and N. M/N are about as old as L3, and are still lacking in Africa (other than relatively recent migrations of M1 and various N lineages). So if L3 really is Eurasian, all of these African L3 lineages would need to have back-migrated shortly after the origin of L3, leaving behind M and N by pure chance, and without leaving a trace of African L3 lineages in Eurasia. Sounds like a fantasy.
DE is a different case, but Y-DNA is very unreliable for tracing demographically significant migrations. Certainly less reliable than mtDNA, at the very least.
Speaking of which, when you included the East African cluster in your previous TreeMix analysis, you found signs of East African gene flow into southern West Eurasians, as well as gene flow from a population most similar to East Africans (which you called "Proto-Eurasian") into the Southwest Asian component. This would explain the increased African affinity of some Eurasians, rather than the back-migration of a very early L3 and/or DE population, unlikely to have been more closely related to certain West Eurasians than to East Eurasians.
Lank, how do you explain the fact that Eurasia has DE and CF, and Africa only E?
ReplyDeleteI mean, if Africa did, indeed have that very high effective population size, and Eurasians are descended from a very small number of Africans, then why did Y chromosomes in Africa get reduced to a single subclade of DE, whereas Eurasia has both DE and CF? We'd expect the opposite to be true: the high size population to experience less drift than the low-size one.
Interestingly, E has been estimated by karafet et al. (2008) as being 52.5ky old. if one fixes CT=70ky. So, how come a newcomer (E) replaces the entire CT clade in Africa but leaves the still older A/B clades in existence? This problem disappears if CT originated in Eurasia.
I think it's much more likely that Y-chromosomes ancestral to CT left Africa pre-100ka, then the population went through a bottleneck in Arabia, c.70ka when the climate deteriorated, and then two founders (CF/DE) survived, the latter re-entering Africa during the crucial 50ka period.
I do agree, however, that East Africans must be further studied as they may hold the key to many questions. The trouble is that they include a triplet of influences from the interior of Africa, Eurasia, and native East African elements, so until we can create "virtual genomes" of their constituent components it is difficult to say much.
ReplyDeleteLank, how do you explain the fact that Eurasia has DE and CF, and Africa only E?
ReplyDeleteI mean, if Africa did, indeed have that very high effective population size, and Eurasians are descended from a very small number of Africans, then why did Y chromosomes in Africa get reduced to a single subclade of DE, whereas Eurasia has both DE and CF? We'd expect the opposite to be true: the high size population to experience less drift than the low-size one.
Interestingly, E has been estimated by karafet et al. (2008) as being 52.5ky old. if one fixes CT=70ky. So, how come a newcomer (E) replaces the entire CT clade in Africa but leaves the still older A/B clades in existence? This problem disappears if CT originated in Eurasia.
I think it's much more likely that Y-chromosomes ancestral to CT left Africa pre-100ka, then the population went through a bottleneck in Arabia, c.70ka when the climate deteriorated, and then two founders (CF/DE) survived, the latter re-entering Africa during the crucial 50ka period.
I have never argued against the clear possibility that DE is of Eurasian origin.
Africans had a higher effective population size, yes. What has happened is that Africa, like Eurasia, was highly affected by recent Y-DNA bottlenecks. Not only are most African males Y-DNA E; most of them also belong to the E1b1 subclade, which is considerably more recent but still managed to replace most other Y chromosomes. E1b1 is really just an East/West African lineage, and more recent expansions to the south (Bantu expansion) and north are the reason for its dominance across the continent. I believe mtDNA is more reliable. L3 originated in East Africa ~70 kya, leading to the OoA bottleneck.
D is East Asian, so DE is certainly not the reason for any increased affinity between West Eurasians and Africans.
I do agree, however, that East Africans must be further studied as they may hold the key to many questions. The trouble is that they include a triplet of influences from the interior of Africa, Eurasia, and native East African elements, so until we can create "virtual genomes" of their constituent components it is difficult to say much.
I agree.
Hi Dienekes,
ReplyDeleteOne thing that might help interpretation is showing the residual plots as well as the trees; this will help you visualize why the algorithm is making the choices it does.
Cheers,
Joe
Africans had a higher effective population size, yes. What has happened is that Africa, like Eurasia, was highly affected by recent Y-DNA bottlenecks.
ReplyDeleteThe point is that Africa is supposed to have a higher effective population size. So, it must have been affected by bottlenecks less than Eurasia was.
We see exactly the opposite: two lineages survive in Eurasia (DE and CF), and one in Africa (DE).
Moreover, if bottlenecks were to blame, and Africa originally had CT, then it's weird that the bottlenecks wiped out the entire CT clade except for E, while at the same time left the A's and B's intact.
"pconroy said... "
ReplyDelete" whenever agriculture spread, it was via male agriculturalists' sons moving to the hunter-gatherer frontier and starting new agricultural communities"
... if Eurasian carrying Y-DNA E moved into the Sahara, then the Sahel region, then West Africa proper, the population would initially be Middle "Eastern in genotype and phenotype, then with demic expansion and some assimilation of the daughters of hunter-gatherers, become more admixed."
"... eventually the male agriculturalist on the "wave of advance" are substantially African by descent, but their Y-DNA is still Middle Eastern or more generally Eurasian."
Most traditional agricultural staples in West Africa were not those of the Near East. They were indigenous and locally domesticated, possibly with multi-millenial periods of semi cultivation first ( cases for the last are most often made for the oil palm and native yam).
Generally speaking speaking there were two zones. The main staples of the sahel/savannah were millet, sorghum and in some places, indigenous rice (oryza glaberrima). The West African forest/Guinea coast region grew the indigenous African yam and semi- cultivated the oil palm.
@Unknown,
ReplyDeleteI also predicted that too in 2010:
http://dienekes.blogspot.com/2010/04/age-and-origin-of-17q21-inversion-in.html
pconroy said...
My take on it would be that it is South West Asia, and from there expanded into Europe and Africa.
Since, to me, agriculture originated in West Asia/Middle East and came in 2 waves to Western Europe:
1. First a faster Coastal Route, around Iberia, and maybe bringing Pastoralists, with sheep/goats
2. Next a slightly later and slower Continental route, like LBK, bringing crop farming and cattle and pigs
Agriculture spread in 2 waves to Africa also:
1. First Pastoralist carrying R1b1 and speaking AfroAsiastic languages - http://en.wikipedia.org/wiki/Afroasiatic_languages - bringing sheep/goats
2. Next crop farming, spread by E1b1b, and spreading into the Sahel, where they are halted by climate for a few thousand years, till they exchange some of their crops for more tropical ones, and spread on again as the Bantu Expansion.
So this would explain the current distribution nicely, and concords well with the evidence.
Wednesday, April 07, 2010 9:35:00 PM
Do these migration edges change the black skeleton of the tree? It did in the other trees but I just can't see whats going on in these trees (I pasted into word and zoomed in).
ReplyDeleteFrom what I CAN see Sardinia and French Basque are NOT showing up near the roots of European branches. They actually seem to show up as ultimate termini, like a recent offshoot of a Kent-like population. This is unexpected.
"The back-migration may have affected all of Africa(...). The main difference between North and Sub-Saharan Africa, is that in the North they found Irhoud-like early H. sapiens, and in Nigeria they found Iwo-Eleru-like archaic forms".
ReplyDeleteThat is not what we see in mtDNA: in North Africa at least 30% of the mtDNA is of likely Iberian origin (not exactly Basque or Sardinian but the arrow did not start in Spain, mind you) and about 75% is of West Eurasian origin. We can see the Iberian and Arabian autosomal components being part of the North African gene pool, etc.
Instead we have never ever seen anything like that in Nigeria. Nothing at all.
Yet the algorithm produces the same result more or less for both, so extremely different, African populations.
I think that it only demonstrates that the program is bugged, that the algorithm is not working as expected and you should report to the designers.
They did warn in the "instructions" paper that some anomalies happened, surely they did not expected anything like this but well...
I see there even genetically so close national names like Norwegians and Swedes separated in own branches and wonder is it sensible to make this kind of trees by preselecting nationalities and in fact branches. There should be some another way to name branches to be faithful for individual level genetic attributes. This used way sounds in some cases like making "nationalistic" history. It is however plausible that genes are much older than nations. The the last millennium has the time of national consolidation, but it obviously has not been enough to build this kind of genetic barriers. I hope that researhers could use national names only as subclassification for GENETIC classification, not for steering genetic results, because using nations as the highest level obviously forces the tool to adapt some distinct genetic attributes. For exmaple of this, Norwegians and Swedes whose national-genetic differences is usually impossible to see at this level.
ReplyDeleteFrom what I CAN see Sardinia and French Basque are NOT showing up near the roots of European branches. They actually seem to show up as ultimate termini, like a recent offshoot of a Kent-like population. This is unexpected.
ReplyDeleteAnnie,
I commented on that in Dienekes' other tree study. Clearly, there is more diversity and a lot of admixture going on in the East in Asia, and the algorithm picks those, first. Without migrations/ admixture on the high-priority list within Europe, it appears that the code prefers a northern population route, there. I am quite sure this will look different in an analysis that concentrates on Europe, with Europe-specific admixtures.
Hopefully, Dienekes has some time and energy left for such a study. ;)
"2. Next crop farming, spread by E1b1b, and spreading into the Sahel, where they are halted by climate for a few thousand years, till they exchange some of their crops for more tropical ones, and spread on again as the Bantu Expansion."
ReplyDeleteMost Berber groups (The more heavily Subsaharan -admixed Tuareg subgroups are the only exceptions I know.) are dominated E1b1b which is also the most common Eurasian subclade. It is relatively rare in Subsaharan Africa and rarer toward the west.
West Africa's most common subclade is E1b1a which occurs at about 90% in the Yoruba and some nearby peoples, near 100% in the proto-Bantu related Bamileke of Cameroon, and is rare in Eurasians
The Near Eastern origin theory for West african farming, though once more popular, is now mostly abandoned by researchers and does not seem to me very likely. Among other reasons, the fact that there is no evidence of overlap or contact between the farming traditions of the two areas during the early phase of the latter makes independent invention seem more plausible.
One might expect some transfer of the Near Eastern package/s of crops, at least to some of the Sahel/savannah, which is much more dry than the forest and not more drastically different from the hotter parts of Western Eurasia that support grain. Agricultural diffusion across eco-zones, especially with the kind of substantial gene flow proposed here, with no evidence of crop transfer at any time(no transitions) is unusual, especially in light of the large environmental ranges gained by Near Eastern cereals, Asian rice, and Maize. Another comparison might be made to the spread of African rice from from the irregularly rainy but often very fertile middle niger floodplain in Mali to the less fertile coastal swamps of Sierra Leone and Senegambia.
The earliest sites in both West African crop zones show a gradual transition to agriculture from a phase of relying on intensive gathering/proto cultivation. This process suggests and is typical of independent domestication and seems less consistent with a local response to the arrival of a fully agricultural group.
Nor is it clear that forest zone farming owed much to the Sahel/Savanah. The earliest dates of both are roughly equal, with tentative evidence of greater age in the first.
Forest crop cultivation also predates the arrival of goats and sheep in the region. Most estimates for earliest oil palm and yam cultivation range from around ca. 4500-2500 bc. in parts of W. Cameroon/S.E .Nigeria. The oldest caprines near the transitional zone are dated 2300-1900 bc in Gajiganna of north Nigeria and 2000-1500 bc in Kintampo of central Ghana. The oldest goats in the S. E Nigeria/W. Cameroon region are ca. 800-500 bc at Yaounde, Cameroon.
I think that in this case Sardinians/Basques are merely acting as a proxy for L3(xM,N)-bearing East Africans. Consider that Yoruba are nearly 100% east African paternally (Y-DNA E) and roughly 1/3 east African on the maternal (L3(xM,N)) side. That should equal roughly 66.5% East African ancestry, which is pretty close to the 64% figure produced in your exercise.
ReplyDelete@Unknown,]
ReplyDeleteIn the above old comments, I should of course have used E1b1, rather than E1b1b.
You have some good points there!
AFAIK, the jury is out on the direction of spread of E1b1a, my take is that it spread in a 3-pronged wave:
1. West to Northern Africa
2. South to Eastern Africa
3. South West to West Africa
All from a source in the Natufian Culture - http://en.wikipedia.org/wiki/Natufian_culture
@Unknown,
ReplyDeletemtDNA L3 is also found in Saudi Arabia, so again I don't think we can definitely say that it evolved in East Africa.
But you are right that it would be informative to see East African, but also North West Africans, Dolgans, Fulbe/Fulani, Socotrans and Yemenis included in a re-analysis.
Dolgan=Dogon
ReplyDeleteRe. the relationship of Sahel farming and early Bantu:
ReplyDeleteThe Bantu expansion involved forest crops and
likely none from the Sahel crops or goats initially. It moved along rivers and sometimes relied a lot on fishing, speeding up later from many factors including the new use of iron. Ancestral Eastern Bantu speakers picked up some of the Sahel package in the E. Central Africa lakes region ca. 600 bc. from Nilo Saharans, and it later spread to some Western Bantu areas. Much Associated vocabulary in said Bantu zones shows Nilo-Saharan or E. Bantu neologistic origins.
"Sardinians/Basques are the only two South-West Eurasian populations included, and any back-migration into Africa must have originated in the southern parts of West Eurasia."
ReplyDeleteMy guess is that the directionality comes from there being more European than African samples, and the Sardinian/Basque link is not particular to those populations but would be general to Southern Europeans who have non-trivial Y-DNA E and mtDNA L, which probably represents secondary expansion Out of Africa in the Holocene, rather than back migration into Africa from Europe.
"how do you explain the fact that Eurasia has DE and CF, and Africa only E?"
ReplyDeleteAfrica has A, B and E, of course. CF is derivative (remotely) from B which is derivative (remotely) from A. DE is also derivative (remotely) from B.
Proto-Eurasians bring Y-DNA CF and mtDNA L3 (or pre-emerged M and N) to Eurasia. DE emerges in Africa, with a handful of martime equipped DE men make their way to Asia (probably after 45kya and before 30kya and close to 30kya than 45kya), have kids with local women in India, and take marginal spaces left over after earlier waves.
Y-DNA E and mtDNA L makes its way by later migration from Africa into Europe in the Holocene.
"DE emerges in Africa, with a handful of martime equipped DE men make their way to Asia (probably after 45kya and before 30kya and close to 30kya than 45kya), have kids with local women in India, and take marginal spaces left over after earlier waves".
ReplyDeleteProblem with that is that D is absent (or virtually so) from India. It is found only in the extreme northeast and there is almost certainly a product of later movement from Tibet/South China. Difficult to make a convincing argument for D having moved easy via India.
"..Problem with that is that D is absent (or virtually so) from India. It is found only in the extreme northeast .."
ReplyDeleteD is also in Andaman islanders. Post neolithic West Eurasian influence is significant even in the south of mainland India, and one imagines more strongly male
>> DE emerges in Africa, with a handful of martime equipped DE men make their way to Asia
ReplyDeleteThose maritime equipped DE men had some ocean-crossing skillz if they made it from Africa to the Andamans.
Actually, there is no reason to think that the expansion of DE was maritime, unless you think Tibet/Mongolia are easily accessible by sea.
">> DE emerges in Africa, with a handful of martime equipped DE men make their way to Asia
ReplyDeleteThose maritime equipped DE men had some ocean-crossing skillz if they made it from Africa to the Andamans.
Actually, there is no reason to think that the expansion of DE was maritime, unless you think Tibet/Mongolia are easily accessible by sea."
Whether it was maritime, went along the S. coast of Asia, or a more inland route, it could have been mostly wiped in certain areas (namely India and much of, now Indo European and Dravidian speaking South Asia) between the Andamans and its current range by the post-neolithic Ancestral North Indian and other West Eurasian migrations.
"D is also in Andaman islanders. Post neolithic West Eurasian influence is significant even in the south of mainland India, and one imagines more strongly male"
ReplyDeleteBut D is absent from South India. And its presence in both the Andamans and in Tibet/South China suggests that the Andamans were settled from somewhere round the northern Bay of Bengal, or possibly from Burma (the closest part of the mainland to the Andamans). Most languages of Burma are classified as 'Tibeto-Burman' so it's reasonable to suspect some influence in Burma from further north. I'm not sure of the extent of haplogroup D in Burma.
This comment has been removed by the author.
ReplyDeleteThis comment has been removed by the author.
ReplyDeleteThis comment has been removed by the author.
ReplyDelete"But D is absent from South India. And its presence in both the Andamans and in Tibet/South China suggests that the Andamans were settled from somewhere round the northern Bay of Bengal, or possibly from Burma (the closest part of the mainland to the Andamans). Most languages of Burma are classified as 'Tibeto-Burman' so it's reasonable to suspect some influence in Burma from further north. I'm not sure of the extent of haplogroup D in Burma."
ReplyDeleteThe Andamanese(with Jarawa at near 100%) carry D*, and Tibeto-Burmans D1. D3a is common in Tibet.
Andamanese languages do not show a relationship to any language family spoken to the northeast, although connections have been suggested to Papuan languages.
Tibeto-Burman was likely spread to the south by neolithic farmers of Mongoloid affinity.
This does not describe the settlement of the Andamans which was much earlier than the neolithic.
Burma was probably, like Thailand and much of south Asia, originally settled by Negrito-like people.
There is an Andamanese-like element (Asi) in South Asia including Northern india and Pakistan and as far north west as the Brahui. Andamanese mtdna M2 and M4 are common in India, unlike Andamanese Ydna haplogroups, which are mostly rare there.
Dieneke, will you be making TreeMix analyses in the near future?
ReplyDelete