Gene. 2012 Jan 31. [Epub ahead of print]
High levels of Paleolithic Y-chromosome lineages characterize Serbia.
Regueiro M, Rivera L, Damnjanovic T, Lukovic L, Milasin J, Herrera RJ.
Abstract
Whether present-day European genetic variation and its distribution patterns can be attributed primarily to the initial peopling of Europe by anatomically modern humans during the Paleolithic, or to latter Near Eastern Neolithic input is still the subject of debate. Southeastern Europe has been a crossroads for several cultures since Paleolithic times and the Balkans, specifically, would have been part of the route used by Neolithic farmers to enter Europe. Given its geographic location in the heart of the Balkan Peninsula at the intersection of Central and Southeastern Europe, Serbia represents a key geographical location that may provide insight to elucidate the interactions between indigenous Paleolithic people and agricultural colonists from the Fertile Crescent. In this study, we examine, for the first time, the Y-chromosome constitution of the general Serbian population. A total of 103 individuals were sampled and their DNA analyzed for 104 Y-chromosome bi-allelic markers and 17 associated STR loci. Our results indicate that approximately 58% of Serbian Y-chromosomes (I1-M253, I2a-P37.2, R1a1a-M198) belong to lineages believed to be pre-Neolithic. On the other hand, the signature of putative Near Eastern Neolithic lineages, including E1b1b1a1-M78, G2a-P15, J1-M267 and J2-M172 and R1b1a2-M269 accounts for 39% of the Y-chromosome. Furthermore, an examination of the distribution of Y-chromosome filiations in Europe indicates extreme levels of Paleolithic lineages in a region encompassing Serbia, Bosnia-Herzegovina and Croatia, possibly the result of Neolithic migrations encroaching on Paleolithic populations against the Adriatic Sea.
Link
February 29, 2012
February 28, 2012
Complete genome of the Tyrolean Iceman
I'll update this post once I read the paper. I could not locate a source for the data after a quick scan of the paper, but:
UPDATE I (Y-chromosome):
From the paper:
In terms of autosomal DNA, the Iceman clearly clusters with modern Sardinians, and also appears slightly more removed than them compared to continental Europeans. Interestingly, at least as far as the PC analyssi shows, Sardinians appear to be intermediate between the Iceman and SW Europeans, rather than Italians. Perhaps, this makes sense if the Paleo-Sardinian language is indeed related to languages of Iberia.
I don't see a downloadable version of the Iceman's genome at the icemangenome.net site, but I've asked the corresponding author for a PLINK/EIGENSOFT version of it. I anticipate that, as I've predicted, this will appear to be largely "Mediterranean" according to Dodecad v3, or "Atlantic_Med" according to the newer K12b calculator. It appears that there has indeed been Sardinian continuity against a backdrop of European discontinuity.
UPDATE III (Sardinians):
The Iceman's genome also places the Sardinian genetic isolate into new light. Two explanations have been proposed for the fact that Sardinians appear genetically distinctive vis a vis continental Europeans:
Nature Communications 3, Article number: 698 doi:10.1038/ncomms1701
New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing
Andreas Keller et al.
The Tyrolean Iceman, a 5,300-year-old Copper age individual, was discovered in 1991 on the Tisenjoch Pass in the Italian part of the Ötztal Alps. Here we report the complete genome sequence of the Iceman and show 100% concordance between the previously reported mitochondrial genome sequence and the consensus sequence generated from our genomic data. We present indications for recent common ancestry between the Iceman and present-day inhabitants of the Tyrrhenian Sea, that the Iceman probably had brown eyes, belonged to blood group O and was lactose intolerant. His genetic predisposition shows an increased risk for coronary heart disease and may have contributed to the development of previously reported vascular calcifications. Sequences corresponding to ~60% of the genome of Borrelia burgdorferi are indicative of the earliest human case of infection with the pathogen for Lyme borreliosis.
Link
After genotyping, we merged both HapMap and 1,000 Genomes genotypes with the Popres/Iceman-merged dataset, resulting in a final analysis dataset containing 125,729 SNPs. PCA was then performed on all samples, excluding the five 1,000 Genomes samples, which were subsequently projected onto the PC space inferred from the rest of the dataset.UPDATE: The Iceman's genome can be found at the http://icemangenome.net site.
UPDATE I (Y-chromosome):
From the paper:
We addressed this issue here by analysing the G2a4-defining L91 SNP in 7,797 chromosomes from 30 regions across Europe. Fig. 3d shows the spatial frequency distribution of G2a4 throughout Europe. The highest frequencies (25 and 9%) occur in southern Corsica and northern Sardinia, respectively, (Fig. 3e) while in mainland Europe the frequencies do not reach 1%.UPDATE II (Autosomal DNA):
In terms of autosomal DNA, the Iceman clearly clusters with modern Sardinians, and also appears slightly more removed than them compared to continental Europeans. Interestingly, at least as far as the PC analyssi shows, Sardinians appear to be intermediate between the Iceman and SW Europeans, rather than Italians. Perhaps, this makes sense if the Paleo-Sardinian language is indeed related to languages of Iberia.
I don't see a downloadable version of the Iceman's genome at the icemangenome.net site, but I've asked the corresponding author for a PLINK/EIGENSOFT version of it. I anticipate that, as I've predicted, this will appear to be largely "Mediterranean" according to Dodecad v3, or "Atlantic_Med" according to the newer K12b calculator. It appears that there has indeed been Sardinian continuity against a backdrop of European discontinuity.
UPDATE III (Sardinians):
The Iceman's genome also places the Sardinian genetic isolate into new light. Two explanations have been proposed for the fact that Sardinians appear genetically distinctive vis a vis continental Europeans:
- Sardinians have lost due to drift alleles that were present in continental Europe
- Continental Europeans have gained alleles that were not present in their Sardinian-like ancestors
Nature Communications 3, Article number: 698 doi:10.1038/ncomms1701
New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing
Andreas Keller et al.
The Tyrolean Iceman, a 5,300-year-old Copper age individual, was discovered in 1991 on the Tisenjoch Pass in the Italian part of the Ötztal Alps. Here we report the complete genome sequence of the Iceman and show 100% concordance between the previously reported mitochondrial genome sequence and the consensus sequence generated from our genomic data. We present indications for recent common ancestry between the Iceman and present-day inhabitants of the Tyrrhenian Sea, that the Iceman probably had brown eyes, belonged to blood group O and was lactose intolerant. His genetic predisposition shows an increased risk for coronary heart disease and may have contributed to the development of previously reported vascular calcifications. Sequences corresponding to ~60% of the genome of Borrelia burgdorferi are indicative of the earliest human case of infection with the pathogen for Lyme borreliosis.
Link
February 25, 2012
Frachetti on the multiregional emergence of mobile pastoralism
 I had previously posted on horses not being important for the emergence of steppe pastoralism, in which Frachetti and Benecke documented how the early (4,500ky) Begash culture of Kazakhstan had developed full-blown pastoralism without apparently relying on horses.
I had previously posted on horses not being important for the emergence of steppe pastoralism, in which Frachetti and Benecke documented how the early (4,500ky) Begash culture of Kazakhstan had developed full-blown pastoralism without apparently relying on horses.This is in contradistinction to both the Botai culture where there is abundant evidence for horse use, apparently for food, as well as the Eneolithic cultures of the European steppe where horse bones are much more prevalent than in early Begash.
The simple model of the emergence of pastoral nomadism proposes the spread of this mode of subsistence from the European (Pontic-Caspian) steppe, together with horses and horse-drawn vehicles. Popularized by David Anthony in recent years, this model views horses and wheels as the great enablers of pastoralism, and views the emergence of pastoral cultures across the steppe as the result of movements of mobile pastoralists -atop their horses, and with their herds- across the Eurasian steppe.
Frachetti is a critic of this model, and proposes instead the importance of the (hitherto neglected) Inner Asian Mountain Corridor as important in facilitating prehistoric contacts between east and west. In a new paper in Current Anthropology he elaborates on his proposed "multiregional" model of the emergence of mobile pastoralism.
I have not read the paper's 38 pages carefully yet (including CA comments and response), but this is clearly a seminal work on the studied topic that will be referenced for years to come. I will post any specific comments in updates to this post. For the moment, I will limit myself to a couple of observations:
- Recent work in Y-chromosome phylogeny has established a fairly disjoint division within haplogroup R1a1a; in particular the R-Z93 subhaplogroup seems to abound in the "Asian steppe", as well as Asia in general, while being generally absent in Europe; the R1a1a and Subclades Y-DNA Project is keeping track of new developments in this field.
- My own research on autosomal DNA, suggests the confluence of two "streams" of ancestry onto the steppe: a west-to-east stream emanating from eastern Europe, and associated with the Atlantic_Baltic (K7b) or North_European (K12b) ancestral component; as well as a West_Asian (K7b) or Caucasus/Gedrosia (K12b) component emanating from the highland regions south of the Caspian and south of the steppe (the traditional Silk Road territory).
In any case, the new paper by Michael Frachetti will provide important new insight to all those who seek to understand "what actually happened" in Eurasian prehistory, and it is a very welcome addition to the ongoing debate.
Current Anthropology Vol. 53, No. 1, February 2012
Multiregional Emergence of Mobile Pastoralism and Nonuniform Institutional Complexity across Eurasia
Michael D. Frachetti
Abstract
In this article I present a new archaeological synthesis concerning the earliest formation of mobile pastoralist economies across central Eurasia. I argue that Eurasian steppe pastoralism developed along distinct local trajectories in the western, central, and (south)eastern steppe, sparking the development of regional networks of interaction in the late fourth and third millennia BC. The “Inner Asian Mountain Corridor” exemplifies the relationship between such incipient regional networks and the process of economic change in the eastern steppe territory. The diverse regional innovations, technologies, and ideologies evident across Eurasia in the mid-third millennium BC are cast as the building blocks of a unique political economy shaped by “nonuniform” institutional alignments among steppe populations throughout the second millennium BC. This theoretical model illustrates how regional channels of interaction between distinct societies positioned Eurasian mobile pastoralists as key players in wide-scale institutional developments among traditionally conceived “core” civilizations while also enabling them to remain strategically independent and small-scale in terms of their own sociopolitical organization. The development of nonuniform institutional complexity among Eurasian pastoralists demonstrates a unique political and economic structure applicable to societies whose variable political and territorial scales are inconsistent with commonly understood evolutionary or corporate sociopolitical typologies such as chiefdoms, states, or empires.
Link
Pre-Neolithic Basque mtDNA gene pool (?)
I haven't read the paper, but I'm unconvinced that an 8,000 YBP estimate of separation (even if it were made with the precision of an atomic clock) "clearly supports" genetic continuity with the Paleolithic/Mesolithic settlers of the Franco-Cantabrian region.
Not only because the date in question is within a millennium or two of the arrival of the Neolithic in Iberia, one of those "curious coincidences."
Much more importantly, because the wisdom of Barbujani continues to be ignored when it comes to tying genetic age estimates to archaeology.
The American Journal of Human Genetics, 23 February 2012 doi:10.1016/j.ajhg.2012.01.002
The Basque Paradigm: Genetic Evidence of a Maternal Continuity in the Franco-Cantabrian Region since Pre-Neolithic Times
Doron M. Behar et al.
Different lines of evidence point to the resettlement of much of western and central Europe by populations from the Franco-Cantabrian region during the Late Glacial and Postglacial periods. In this context, the study of the genetic diversity of contemporary Basques, a population located at the epicenter of the Franco-Cantabrian region, is particularly useful because they speak a non-Indo-European language that is considered to be a linguistic isolate. In contrast with genome-wide analysis and Y chromosome data, where the problem of poor time estimates remains, a new timescale has been established for the human mtDNA and makes this genome the most informative marker for studying European prehistory. Here, we aim to increase knowledge of the origins of the Basque people and, more generally, of the role of the Franco-Cantabrian refuge in the postglacial repopulation of Europe. We thus characterize the maternal ancestry of 908 Basque and non-Basque individuals from the Basque Country and immediate adjacent regions and, by sequencing 420 complete mtDNA genomes, we focused on haplogroup H. We identified six mtDNA haplogroups, H1j1, H1t1, H2a5a1, H1av1, H3c2a, and H1e1a1, which are autochthonous to the Franco-Cantabrian region and, more specifically, to Basque-speaking populations. We detected signals of the expansion of these haplogroups at ∼4,000 years before present (YBP) and estimated their separation from the pan-European gene pool at ∼8,000 YBP, antedating the Indo-European arrival to the region. Our results clearly support the hypothesis of a partial genetic continuity of contemporary Basques with the preceding Paleolithic/Mesolithic settlers of their homeland.
Link
Not only because the date in question is within a millennium or two of the arrival of the Neolithic in Iberia, one of those "curious coincidences."
Much more importantly, because the wisdom of Barbujani continues to be ignored when it comes to tying genetic age estimates to archaeology.
The American Journal of Human Genetics, 23 February 2012 doi:10.1016/j.ajhg.2012.01.002
The Basque Paradigm: Genetic Evidence of a Maternal Continuity in the Franco-Cantabrian Region since Pre-Neolithic Times
Doron M. Behar et al.
Different lines of evidence point to the resettlement of much of western and central Europe by populations from the Franco-Cantabrian region during the Late Glacial and Postglacial periods. In this context, the study of the genetic diversity of contemporary Basques, a population located at the epicenter of the Franco-Cantabrian region, is particularly useful because they speak a non-Indo-European language that is considered to be a linguistic isolate. In contrast with genome-wide analysis and Y chromosome data, where the problem of poor time estimates remains, a new timescale has been established for the human mtDNA and makes this genome the most informative marker for studying European prehistory. Here, we aim to increase knowledge of the origins of the Basque people and, more generally, of the role of the Franco-Cantabrian refuge in the postglacial repopulation of Europe. We thus characterize the maternal ancestry of 908 Basque and non-Basque individuals from the Basque Country and immediate adjacent regions and, by sequencing 420 complete mtDNA genomes, we focused on haplogroup H. We identified six mtDNA haplogroups, H1j1, H1t1, H2a5a1, H1av1, H3c2a, and H1e1a1, which are autochthonous to the Franco-Cantabrian region and, more specifically, to Basque-speaking populations. We detected signals of the expansion of these haplogroups at ∼4,000 years before present (YBP) and estimated their separation from the pan-European gene pool at ∼8,000 YBP, antedating the Indo-European arrival to the region. Our results clearly support the hypothesis of a partial genetic continuity of contemporary Basques with the preceding Paleolithic/Mesolithic settlers of their homeland.
Link
February 23, 2012
Lack of rain led to collapse of Maya civilization
I bet that the 2012-Mayan-calendar-end-of-the-world-as-we-know-it crowd will proceed undeterred, at least until December, to ascribe preternatural powers of prediction of the far future to the Maya, just as science tells us that they failed to adjust to a relatively mild ecological shift during their own time.
From the press release:
"Our results show rather modest rainfall reductions between times when the Classic Maya Civilization flourished and its collapse – between AD 800-950. These reductions amount to only 25 to 40 per cent in annual rainfall. But they were large enough for evaporation to become dominant over rainfall, and open water availability was rapidly reduced. The data suggest that the main cause was a decrease in summer storm activity."
Science 24 February 2012:Vol. 335 no. 6071 pp. 956-959 DOI: 10.1126/science.1216629
Collapse of Classic Maya Civilization Related to Modest Reduction in Precipitation
Martín Medina-Elizalde, Eelco J. Rohling
The disintegration of the Classic Maya civilization in the Yucatán Peninsula and Central America was a complex process that occurred over an approximately 200-year interval and involved a catastrophic depopulation of the region. Although it is well established that the civilization collapse coincided with widespread episodes of drought, their nature and severity remain enigmatic. We present a quantitative analysis that offers a coherent interpretation of four of the most detailed paleoclimate records of the event. We conclude that the droughts occurring during the disintegration of the Maya civilization represented up to a 40% reduction in annual precipitation, probably due to a reduction in summer season tropical storm frequency and intensity.
More on 43,000-year old "Neanderthal" art
I had blogged about this based on an early story. There is now an article in the far more reputable New Scientist which puts it into proper perspective:
Looking oddly akin to the DNA double helix, the images in fact depict the seals that the locals would have eaten, says José Luis Sanchidrián at the University of Cordoba, Spain. They have "no parallel in Palaeolithic art", he adds. His team say that charcoal remains found beside six of the paintings – preserved in Spain's Nerja caves – have been radiocarbon dated to between 43,500 and 42,300 years old.
...
If the age is confirmed, Pettitt suggests that the cave paintings could still have been the work of modern humans. "We can't be absolutely sure that Homo sapiens were not down there in the south of Spain at this time," he says.
Sanchidrián does not rule out the possibility that the paintings were made by early Homo sapiens but says that this theory is "much more hypothetical" than the idea that Neanderthals were behind them.
Dating of the Nerja seal paintings' pigments will not take place until after 2013. Further excavations in the extensive cave system – discovered by a group of boys hunting bats in 1959 – is ongoing.It will be interesting to see the dates from the pigments themselves. As of now, I would rank the possibilities of who made this art, from most to least probable as: modern humans long after 43,000 years ago; modern humans c. 43,000 years ago; Neandertals c. 43,000 years ago.
February 22, 2012
ChromoPainter/fineSTRUCTURE analysis of select South Asian/West Eurasian populations
This is the final result of the analysis mentioned in this previous post on the Kalash, using all 22 chromosomes.
Due to the quadratic running time of ChromoPainter, I took a random sample of 15 individuals from every included population with more than15 individuals. The final set included 392 individuals. It appears that a set of ~400 individuals/~260k SNPs can be processed in about 2 weeks on a single thread.
The raw chunkcounts between all individuals can be obtained from here.
The heatmap can be seen below:
The principal components analysis, shows the familiar West-to-South Asia cline:
More information can be found in the spreadsheet, including:
Finally, in the RAR file you can find some plots of Z scores (by row) for the different population.
For example, here is a list of donors for the Kalash population; the order is slightly different compared to the teaser, but the overall pattern is the same.
Compare with an outbred population, such as the Armenians:
Due to the quadratic running time of ChromoPainter, I took a random sample of 15 individuals from every included population with more than15 individuals. The final set included 392 individuals. It appears that a set of ~400 individuals/~260k SNPs can be processed in about 2 weeks on a single thread.
The raw chunkcounts between all individuals can be obtained from here.
The heatmap can be seen below:
The principal components analysis, shows the familiar West-to-South Asia cline:
More information can be found in the spreadsheet, including:
- How many individuals from each population were assigned to each of 51 clusters
- Individual assignments of all 392 individuals
- Raw chunkcounts between all 33 different populations
- Z scores of the above (by row)
- Z scores of the above (by column)
- by row: scan each line to see which populations (columns) are the bigger donors for each row.
- by column: scan each column to see which populations (rows) are the bigger recipients for each column.
Finally, in the RAR file you can find some plots of Z scores (by row) for the different population.
For example, here is a list of donors for the Kalash population; the order is slightly different compared to the teaser, but the overall pattern is the same.
February 18, 2012
A teaser on the Kalash
UPDATE (Feb 22): The complete analysis can be found here.
Razib has a post on Kalash on the human tree. As it happens, I am in the middle of a ChromoPainter/fineSTRUCTURE analysis of a broad dataset designed to explore certain mysteries that have often come up in my previous experiments. Barring the unexpected, the analysis should be completed sometime next week.
Below you can see the normalized number of "chunks" donated by various populations to the Kalash. First, we normalize including intra-Kalash sharing:
Notice the extreme intra-Kalash haplotype sharing: Kalash individuals are recipient of "chunks" from other Kalash individuals ~5 standard deviations more often than the mean over this set of populations.
However, if we igonore intra-Kalash haplotype sharing, then the donor populations are:
Of particular interest is the fact that all West Asian populations appear higher on the donor list than all Northern European ones, which confirms, using a haplotype-based approach, my previous inference that the Ancestral North Indian (ANI) component is related to West Asians.
Razib has a post on Kalash on the human tree. As it happens, I am in the middle of a ChromoPainter/fineSTRUCTURE analysis of a broad dataset designed to explore certain mysteries that have often come up in my previous experiments. Barring the unexpected, the analysis should be completed sometime next week.
Below you can see the normalized number of "chunks" donated by various populations to the Kalash. First, we normalize including intra-Kalash sharing:
Notice the extreme intra-Kalash haplotype sharing: Kalash individuals are recipient of "chunks" from other Kalash individuals ~5 standard deviations more often than the mean over this set of populations.
However, if we igonore intra-Kalash haplotype sharing, then the donor populations are:
Of particular interest is the fact that all West Asian populations appear higher on the donor list than all Northern European ones, which confirms, using a haplotype-based approach, my previous inference that the Ancestral North Indian (ANI) component is related to West Asians.
February 17, 2012
Adaptation to high altitude in Ethiopian highlands
There was another paper on adaptation to high altitude of Tibetans.
Another article on Tibetans and Andean high-altitude adaptation, and another one by Bigham et al.
Genome Biology 2012, 13:R1 doi:10.1186/gb-2012-13-1-r1
Genetic adaptation to high altitude in the Ethiopian highlands
Laura B Scheinfeldt et al.
Abstract (provisional)
Background
Genomic analysis of high-altitude populations residing in the Andes and Tibet has revealed several candidate loci for involvement in high-altitude adaptation, a subset of which have also been shown to be associated with hemoglobin levels, including EPAS1, EGLN1, and PPARA, which play a role in the HIF-1 pathway. Here, we have extended this work to high and low altitude populations living in Ethiopia for which we have measured hemoglobin levels. We genotyped the Illumina 1M SNP array and employed several genome-wide scans for selection and targeted association with hemoglobin levels to identify genes that play a role in adaptation to high altitude.
Results
We have identified a set of candidate genes for positive selection in our high-altitude population sample, demonstrated significantly different hemoglobin levels between high and low altitude Ethiopians and have identified a subset of candidate genes for selection, several of which also show suggestive associations with hemoglobin levels.
Conclusions
We highlight several candidate genes for involvement in high-altitude adaptation in Ethiopia, including CBARA1, VAV3, ARNT2 and THRB. Although most of these genes have not been identified in previous studies of high-altitude Tibetan or Andean population samples, two of these genes (THRB and ARNT2) play a role in the HIF-1 pathway, a pathway implicated in previous work reported in Tibetan and Andean studies. These combined results suggest that adaptation to high altitude arose independently due to convergent evolution in high-altitude Amhara populations in Ethiopia.
Link
Another article on Tibetans and Andean high-altitude adaptation, and another one by Bigham et al.
Genome Biology 2012, 13:R1 doi:10.1186/gb-2012-13-1-r1
Genetic adaptation to high altitude in the Ethiopian highlands
Laura B Scheinfeldt et al.
Abstract (provisional)
Background
Genomic analysis of high-altitude populations residing in the Andes and Tibet has revealed several candidate loci for involvement in high-altitude adaptation, a subset of which have also been shown to be associated with hemoglobin levels, including EPAS1, EGLN1, and PPARA, which play a role in the HIF-1 pathway. Here, we have extended this work to high and low altitude populations living in Ethiopia for which we have measured hemoglobin levels. We genotyped the Illumina 1M SNP array and employed several genome-wide scans for selection and targeted association with hemoglobin levels to identify genes that play a role in adaptation to high altitude.
Results
We have identified a set of candidate genes for positive selection in our high-altitude population sample, demonstrated significantly different hemoglobin levels between high and low altitude Ethiopians and have identified a subset of candidate genes for selection, several of which also show suggestive associations with hemoglobin levels.
Conclusions
We highlight several candidate genes for involvement in high-altitude adaptation in Ethiopia, including CBARA1, VAV3, ARNT2 and THRB. Although most of these genes have not been identified in previous studies of high-altitude Tibetan or Andean population samples, two of these genes (THRB and ARNT2) play a role in the HIF-1 pathway, a pathway implicated in previous work reported in Tibetan and Andean studies. These combined results suggest that adaptation to high altitude arose independently due to convergent evolution in high-altitude Amhara populations in Ethiopia.
Link
February 16, 2012
First look at Turkish and Kyrgyz data from Hodoğlugil & Mahley (2012)
The authors of the recent paper on Turkish population structure were kind enough to share their data with me. I will be sure to use this data in future experiments, such as the ChromoPainter and fastIBD analysis of Balkans/West Asia, as well as a ChromoPainter analysis of Altaic speakers, following on the footsteps of my recent analysis of Afroasiatic speakers.
PCA
As a first step, after processing the new data, I carried out a PCA analysis (in smartpca with no outlier removal iterations), combined with various Turkic groups, as well as a few neighbors of Anatolian Turks, combining data from the literature and the Dodecad Project.
The Turkic cline from East to West Eurasia, observed by myself and others in various experiments is again evident.
The blowup of the above, focusing on the West Eurasian portion (top right) is easier to read:
As always, population labels are placed in the average position of each population. So, for example, the Behar et al. Iranians_19 sample is shifted to the left, because of the existence of a few African admixed individuals in this sample. The Iranian_D sample of Project participants seem to lack this admixture.
Also, note that since there is no South Asian reference in this first experiment, Iranians overlap with Turks along the first two dimensions. As we've seen in the Dodecad Project, both Iranians and Anatolian Turks are "eastward-shifted" relative to other West Eurasians, but the former have a strong South Asian- and the latter a Central Asian- tendency.
The new Kyrgyz sample falls between the Kazakh and the Altai along the cline, and is more "eastern" compared to the Uygurs and Uzbeks, and more "western" compared to Altai, Tuva, and Dolgans.
Kayseri and Istanbul Turks overlap with Behar et al. Turks as well as the Turkish_D sample. The Aydin sample appears to be more heterogenous, with a more eastern overall center of weight. More on this below.
ADMIXTURE
I also carried out a K=3 ADMIXTURE analysis of the dataset.
Below are the population portraits for the three new Turkish samples, as well as the Kyrgyz sample:
It is obvious that many Turks have low levels of Asian admixture, lacking in their geographical neighbors, but this is quite variable on an individual basis.
UPDATE (17 Feb):
I have also assessed the new data with the K12b calculator. Below are the normalized median proportions.
PCA
As a first step, after processing the new data, I carried out a PCA analysis (in smartpca with no outlier removal iterations), combined with various Turkic groups, as well as a few neighbors of Anatolian Turks, combining data from the literature and the Dodecad Project.
The Turkic cline from East to West Eurasia, observed by myself and others in various experiments is again evident.
The blowup of the above, focusing on the West Eurasian portion (top right) is easier to read:
As always, population labels are placed in the average position of each population. So, for example, the Behar et al. Iranians_19 sample is shifted to the left, because of the existence of a few African admixed individuals in this sample. The Iranian_D sample of Project participants seem to lack this admixture.
Also, note that since there is no South Asian reference in this first experiment, Iranians overlap with Turks along the first two dimensions. As we've seen in the Dodecad Project, both Iranians and Anatolian Turks are "eastward-shifted" relative to other West Eurasians, but the former have a strong South Asian- and the latter a Central Asian- tendency.
The new Kyrgyz sample falls between the Kazakh and the Altai along the cline, and is more "eastern" compared to the Uygurs and Uzbeks, and more "western" compared to Altai, Tuva, and Dolgans.
Kayseri and Istanbul Turks overlap with Behar et al. Turks as well as the Turkish_D sample. The Aydin sample appears to be more heterogenous, with a more eastern overall center of weight. More on this below.
ADMIXTURE
I also carried out a K=3 ADMIXTURE analysis of the dataset.
Below are the population portraits for the three new Turkish samples, as well as the Kyrgyz sample:
UPDATE (17 Feb):
I have also assessed the new data with the K12b calculator. Below are the normalized median proportions.
More criticism of language origin in Southwest Africa paper (Hunley et al. 2012)
The second co-author, Claire Bowern, writes to me:
Another recent paper by Cysouw et al. here, also critical of the Atkinson (2011) paper.
Proceedings of the Royal Society B doi: 10.1098/rspb.2011.2296
Rejection of a serial founder effects model of genetic and linguistic coevolution
Keith Hunley et al.
Recent genetic studies attribute the negative correlation between population genetic diversity and distance from Africa to a serial founder effects (SFE) evolutionary process. A recent linguistic study concluded that a similar decay in phoneme inventories in human languages was also the product of the SFE process. However, the SFE process makes additional predictions for patterns of neutral genetic diversity, both within and between groups, that have not yet been tested on phonemic data. In this study, we describe these predictions and test them on linguistic and genetic samples. The linguistic sample consists of 725 widespread languages, which together contain 908 distinct phonemes. The genetic sample consists of 614 autosomal microsatellite loci in 100 widespread populations. All aspects of the genetic pattern are consistent with the predictions of SFE. In contrast, most of the predictions of SFE are violated for the phonemic data. We show that phoneme inventories provide information about recent contacts between languages. However, because phonemes change rapidly, they cannot provide information about more ancient evolutionary processes.
Link
Language is great for looking at the Holocene, but it changes too fast for it to be useful for more remote relations. The point is not to question the origin of language in Africa, but to question the utility of modern linguistic data for providing answers to such questions.From the paper:
we find that the correlation between phoneme levels and distance from putative origins is most negative when the origin is located in Eurasia, not Africa (figure 3d), implying that phonemic diversity has not been moulded at the global level by the same evolutionary processes that shaped neutral genetic diversity.The paper also has free supplementary material here, from which the tree of Indo-European languages (left) is taken. The numbers attached to languages indicate the number of vowels/consonants in their phonemic inventory.
Also contrary to the predictions of SFE, the correlation between phoneme inventory size and geographical distance was most positive when the origin was located in Oceania rather than in the Americas. This is because there is a relative deficit of total and private phonemes in Oceania, and an excess in the Americas.
...
The non-tree-like pattern of phonemic variation is also inconsistent with the predictions of SFE. Because we were unable to construct a robust phoneme tree, we are unable to determine whether the observed correlations between phonemic difference and geographical distance are a by-product of the SFE process or the result of phonemic exchange between neighbouring languages. The fact that the correlations exist within regions independent of language family status, however, indicates that local exchange is responsible for at least some of the correlation.
...
Having diversified within the last 10 000 years, currently attested language families are young relative to the age of our species, and specialists have had success reconstructing the evolutionary process in many of them [3,24–28]. Only the IE correlation reached statistical significance, but the correlation was positive.
Another recent paper by Cysouw et al. here, also critical of the Atkinson (2011) paper.
Proceedings of the Royal Society B doi: 10.1098/rspb.2011.2296
Rejection of a serial founder effects model of genetic and linguistic coevolution
Keith Hunley et al.
Recent genetic studies attribute the negative correlation between population genetic diversity and distance from Africa to a serial founder effects (SFE) evolutionary process. A recent linguistic study concluded that a similar decay in phoneme inventories in human languages was also the product of the SFE process. However, the SFE process makes additional predictions for patterns of neutral genetic diversity, both within and between groups, that have not yet been tested on phonemic data. In this study, we describe these predictions and test them on linguistic and genetic samples. The linguistic sample consists of 725 widespread languages, which together contain 908 distinct phonemes. The genetic sample consists of 614 autosomal microsatellite loci in 100 widespread populations. All aspects of the genetic pattern are consistent with the predictions of SFE. In contrast, most of the predictions of SFE are violated for the phonemic data. We show that phoneme inventories provide information about recent contacts between languages. However, because phonemes change rapidly, they cannot provide information about more ancient evolutionary processes.
Link
February 15, 2012
Origin of language in Africa questioned
A new paper in Science is criticial of another recent paper in the same journal, which proposed that there was a diminution of phonemic diversity away from Southwest Africa, suggesting that region as the "birthplace of language".
I had expressed scepticism of that paper on different grounds; also somewhat related, I recently chanced upon a piece in Scientific American suggests that click sounds, for which languages of South Africa are famous are also found in English.
Getting back to the current article, quoting from a preprint I had obtained a while back:
It would be great if linguistic methods could pinpoint the origin of language, but I am not so sure that it can be done. We don't even know which groups of hominins could speak. Interestingly a Kebaran Neandertal apparently had hyoid bones that suggested a modern-like capacity for speech. On the other hand, Kebara was apparently atypical of Neandertals in other aspects of morphology, deviating in a modern human direction, and postdating the major human expansion placed c. 70,000 years ago.
So, it may very well be that language was indeed unique to Homo sapiens, although, as the current paper aptly shows, tracing its origin in space may prove to be quite a challenge.
Science 10 February 2012: Vol. 335 no. 6069 p. 657 DOI: 10.1126/science.1208841
Comment on “Phonemic Diversity Supports a Serial Founder Effect Model of Language Expansion from Africa”
Michael Cysouw et al.
We show that Atkinson’s (Reports, 15 April 2011, p. 346) intriguing proposal—that global linguistic diversity supports a single language origin in Africa—is an artifact of using suboptimal data, biased methodology, and unjustified assumptions. We criticize his approach using more suitable data, and we additionally provide new results suggesting a more complex scenario for the emergence of global linguistic diversity.
Link
I had expressed scepticism of that paper on different grounds; also somewhat related, I recently chanced upon a piece in Scientific American suggests that click sounds, for which languages of South Africa are famous are also found in English.
Getting back to the current article, quoting from a preprint I had obtained a while back:
Notwithstanding this criticism, we replicated Atkinson’s method using the UPSID data, but instead of a single origin in West Africa, we found two separate ‘origins’, one in East Africa and one in the Caucasus (Fig. S10). The BIC+4 range of possible origins covers a large area including also the Middle East and South Africa. Although this finding does not necessarily contradict an expansion form Africa, it does not provide clear support in its favor, either. Further, adding a quadratic distance factor to the model substantially improves the fit and suggests an alternative origin located in New Guinea with a small phoneme inventory (Fig. S10). Even more problematic, when we apply the original method to other inventory-like linguistic characteristics from WALS (Fig. 1) we find origins of global clines all over the world, not just in Africa, and not always corresponding to the highest structural ‘complexity’ (Fig. S11). Therefore, the observation of an Africa-based phoneme inventory cline does not generalize to other linguistic characteristics of a similar kind.
It would be great if linguistic methods could pinpoint the origin of language, but I am not so sure that it can be done. We don't even know which groups of hominins could speak. Interestingly a Kebaran Neandertal apparently had hyoid bones that suggested a modern-like capacity for speech. On the other hand, Kebara was apparently atypical of Neandertals in other aspects of morphology, deviating in a modern human direction, and postdating the major human expansion placed c. 70,000 years ago.
So, it may very well be that language was indeed unique to Homo sapiens, although, as the current paper aptly shows, tracing its origin in space may prove to be quite a challenge.
Science 10 February 2012: Vol. 335 no. 6069 p. 657 DOI: 10.1126/science.1208841
Comment on “Phonemic Diversity Supports a Serial Founder Effect Model of Language Expansion from Africa”
Michael Cysouw et al.
We show that Atkinson’s (Reports, 15 April 2011, p. 346) intriguing proposal—that global linguistic diversity supports a single language origin in Africa—is an artifact of using suboptimal data, biased methodology, and unjustified assumptions. We criticize his approach using more suitable data, and we additionally provide new results suggesting a more complex scenario for the emergence of global linguistic diversity.
Link
Turkish population structure (Hodoğlugil and Mahley, 2012)
These results appear to be consistent with my own estimate of 1/7 Central Asian ancestry for Anatolian Turks. In a more recent analysis based on haplotype sharing, I have discovered that there is some sub-structure within the Turkish population, with the emergence of three tendencies: a central group of Anatolian Turks, a group of Turks with partial origin from the Balkans, and a group of Turks from northeastern Anatolia. As more project participants join the Dodecad Project, we will learn much more on the structure of the Turkish population and its relationships with its geographical neighbors and linguistic relatives.
Annals of Human Genetics DOI: 10.1111/j.1469-1809.2011.00701.x
Turkish Population Structure and Genetic Ancestry Reveal Relatedness among Eurasian Populations
Uğur Hodoğlugil, Robert W. Mahley
Turkey has experienced major population movements. Population structure and genetic relatedness of samples from three regions of Turkey, using over 500,000 SNP genotypes, were compared together with Human Genome Diversity Panel (HGDP) data. To obtain a more representative sampling from Central Asia, Kyrgyz samples (Bishkek, Kyrgyzstan) were genotyped and analysed. Principal component (PC) analysis reveals a significant overlap between Turks and Middle Easterners and a relationship with Europeans and South and Central Asians; however, the Turkish genetic structure is unique. FRAPPE, STRUCTURE, and phylogenetic analyses support the PC analysis depending upon the number of parental ancestry components chosen. For example, supervised STRUCTURE (K= 3) illustrates a genetic ancestry for the Turks of 45% Middle Eastern (95% CI, 42–49), 40% European (95% CI, 36–44) and 15% Central Asian (95% CI, 13–16), whereas at K= 4 the genetic ancestry of the Turks was 38% European (95% CI, 35–42), 35% Middle Eastern (95% CI, 33–38), 18% South Asian (95% CI, 16–19) and 9% Central Asian (95% CI, 7–11). PC analysis and FRAPPE/STRUCTURE results from three regions in Turkey (Aydin, Istanbul and Kayseri) were superimposed, without clear subpopulation structure, suggesting sample homogeneity. Thus, this study demonstrates admixture of Turkish people reflecting the population migration patterns.
Link
Annals of Human Genetics DOI: 10.1111/j.1469-1809.2011.00701.x
Turkish Population Structure and Genetic Ancestry Reveal Relatedness among Eurasian Populations
Uğur Hodoğlugil, Robert W. Mahley
Turkey has experienced major population movements. Population structure and genetic relatedness of samples from three regions of Turkey, using over 500,000 SNP genotypes, were compared together with Human Genome Diversity Panel (HGDP) data. To obtain a more representative sampling from Central Asia, Kyrgyz samples (Bishkek, Kyrgyzstan) were genotyped and analysed. Principal component (PC) analysis reveals a significant overlap between Turks and Middle Easterners and a relationship with Europeans and South and Central Asians; however, the Turkish genetic structure is unique. FRAPPE, STRUCTURE, and phylogenetic analyses support the PC analysis depending upon the number of parental ancestry components chosen. For example, supervised STRUCTURE (K= 3) illustrates a genetic ancestry for the Turks of 45% Middle Eastern (95% CI, 42–49), 40% European (95% CI, 36–44) and 15% Central Asian (95% CI, 13–16), whereas at K= 4 the genetic ancestry of the Turks was 38% European (95% CI, 35–42), 35% Middle Eastern (95% CI, 33–38), 18% South Asian (95% CI, 16–19) and 9% Central Asian (95% CI, 7–11). PC analysis and FRAPPE/STRUCTURE results from three regions in Turkey (Aydin, Istanbul and Kayseri) were superimposed, without clear subpopulation structure, suggesting sample homogeneity. Thus, this study demonstrates admixture of Turkish people reflecting the population migration patterns.
Link
Independent domestication of rice varieties
Mol Biol Evol (2011) doi: 10.1093/molbev/msr315
Independent Domestication of Asian Rice Followed by Gene Flow from japonica to indica
Ching-chia Yang et al.
Results from studies on the domestication process of Asian rice Oryza sativa have been controversial because of its complicated evolutionary history. Previous studies have yielded two alternative hypotheses about the origin(s) of the two major groups of O. sativa: japonica and indica. One study proposes a single common wild ancestor, whereas the other suggests that there were multiple domestication events of different types of wild rice. Here, we provide clear evidence of the independent domestication of japonica and indica obtained via high-throughput sequencing and a large-scale comparative analysis of two wild rice accessions (W1943 and W0106) and two cultivars (a japonica cultivar called “Nipponbare” and an indica cultivar called “Guangluai-4”). The different domestication processes of the two cultivar groups appear to have led to distinct patterns of molecular evolution in protein-coding regions. The intensity of purifying selection was relaxed only in the japonica group, possibly because of a bottleneck effect. Moreover, a genome-wide comparison between Nipponbare, Guangluai-4, and another indica cultivar (93-11) suggests multiple hybridization events between japonica and indica, both before and after the divergence of the indica cultivars. We found that a large amount of genomic DNA, including domestication-related genes, was transferred from japonica to indica, which might have been important in the development of modern rice. Our study provides an overview of the dynamic process of Asian rice domestication, including independent domestication events and subsequent gene flow.
Link
Independent Domestication of Asian Rice Followed by Gene Flow from japonica to indica
Ching-chia Yang et al.
Results from studies on the domestication process of Asian rice Oryza sativa have been controversial because of its complicated evolutionary history. Previous studies have yielded two alternative hypotheses about the origin(s) of the two major groups of O. sativa: japonica and indica. One study proposes a single common wild ancestor, whereas the other suggests that there were multiple domestication events of different types of wild rice. Here, we provide clear evidence of the independent domestication of japonica and indica obtained via high-throughput sequencing and a large-scale comparative analysis of two wild rice accessions (W1943 and W0106) and two cultivars (a japonica cultivar called “Nipponbare” and an indica cultivar called “Guangluai-4”). The different domestication processes of the two cultivar groups appear to have led to distinct patterns of molecular evolution in protein-coding regions. The intensity of purifying selection was relaxed only in the japonica group, possibly because of a bottleneck effect. Moreover, a genome-wide comparison between Nipponbare, Guangluai-4, and another indica cultivar (93-11) suggests multiple hybridization events between japonica and indica, both before and after the divergence of the indica cultivars. We found that a large amount of genomic DNA, including domestication-related genes, was transferred from japonica to indica, which might have been important in the development of modern rice. Our study provides an overview of the dynamic process of Asian rice domestication, including independent domestication events and subsequent gene flow.
Link
February 11, 2012
February 10, 2012
Facial attractiveness and interracial marriage
The data:
From the paper:
So, what this study shows is that while intra-racial marriage is still the norm, black males have an easier time overcoming the racial barrier compared to black females, and Asian females compared to Asian males.
The paper does not seem to present any facial attractiveness data, although it does present an analysis to discount an alternative hypothesis based on stature differences. According to that hypothesis, Asian females outmarry more easily than Asian males because Asians are shorter, and women tend to marry taller men than themselves.
PLoS ONE 7(2): e31703. doi:10.1371/journal.pone.0031703
A Facial Attractiveness Account of Gender Asymmetries in Interracial Marriage
Michael B. Lewis
Abstract
Background
In the US and UK, more Black men are married to White women than vice versa and there are more White men married to Asian women than vice versa. Models of interracial marriage, based on the exchange of racial status for other capital, cannot explain these asymmetries. A new explanation is offered based on the relative perceived facial attractiveness of the different race-by-gender groups.
Method and Findings
This explanation was tested using a survey of perceived facial attractiveness. This found that Black males are perceived as more attractive than White or East Asian males whereas among females, it is the East Asians that are perceived as most attractive on average.
Conclusions
Incorporating these attractiveness patterns into the model of marriage decisions produces asymmetries in interracial marriage similar to those in the observed data in terms of direction and relative size. This model does not require differences in status between races nor different strategies based on gender. Predictions are also generated regarding the relative attractiveness of those engaging in interracial marriage.
Link
From the paper:
The results of the experiment demonstrated that there are robust differences in the relative perceived attractiveness of different racial groups. Further, these differences are affected by the gender of the person being rated. Among males, Black faces were rated as the most attractive followed by White faces and then Asian faces. For the females, Asian faces were seen as the most attractive followed by White and then Black faces. The same pattern was found regardless of the ethnicity of the person doing the ratings.Whatever the perceptions of attractiveness, it seems that people still tend to marry within their own races. For example, in the UK 0.24% of white females marry black males, and the corresponding percentage for the US is 0.56%. Since blacks make up roughly 1/10 of the population in the US, then if race was not an issue, we'd expect white females to marry black males about 10% of the time; the empirical figure is about ~20 times lower.
So, what this study shows is that while intra-racial marriage is still the norm, black males have an easier time overcoming the racial barrier compared to black females, and Asian females compared to Asian males.
The paper does not seem to present any facial attractiveness data, although it does present an analysis to discount an alternative hypothesis based on stature differences. According to that hypothesis, Asian females outmarry more easily than Asian males because Asians are shorter, and women tend to marry taller men than themselves.
PLoS ONE 7(2): e31703. doi:10.1371/journal.pone.0031703
A Facial Attractiveness Account of Gender Asymmetries in Interracial Marriage
Michael B. Lewis
Abstract
Background
In the US and UK, more Black men are married to White women than vice versa and there are more White men married to Asian women than vice versa. Models of interracial marriage, based on the exchange of racial status for other capital, cannot explain these asymmetries. A new explanation is offered based on the relative perceived facial attractiveness of the different race-by-gender groups.
Method and Findings
This explanation was tested using a survey of perceived facial attractiveness. This found that Black males are perceived as more attractive than White or East Asian males whereas among females, it is the East Asians that are perceived as most attractive on average.
Conclusions
Incorporating these attractiveness patterns into the model of marriage decisions produces asymmetries in interracial marriage similar to those in the observed data in terms of direction and relative size. This model does not require differences in status between races nor different strategies based on gender. Predictions are also generated regarding the relative attractiveness of those engaging in interracial marriage.
Link
February 09, 2012
Y-chromosomes and coronary artery disease in Britain
I first reported about this research in 2010. Now, a paper on the topic has appeared in The Lancet. Apart from the potential health implications, it serves to remind us that at least some of the dramatic shifts in Y-haplogroup frequencies between modern and ancient populations may have been driven -at least in part- by natural selection.
The Lancet, Early Online Publication, 9 February 2012 doi:10.1016/S0140-6736(11)61453-0
Inheritance of coronary artery disease in men: an analysis of the role of the Y chromosome
Fadi J Charchar PhD et al.
Summary
Background A sexual dimorphism exists in the incidence and prevalence of coronary artery disease—men are more commonly affected than are age-matched women. We explored the role of the Y chromosome in coronary artery disease in the context of this sexual inequity.
Methods We genotyped 11 markers of the male-specific region of the Y chromosome in 3233 biologically unrelated British men from three cohorts: the British Heart Foundation Family Heart Study (BHF-FHS), West of Scotland Coronary Prevention Study (WOSCOPS), and Cardiogenics Study. On the basis of this information, each Y chromosome was tracked back into one of 13 ancient lineages defined as haplogroups. We then examined associations between common Y chromosome haplogroups and the risk of coronary artery disease in cross-sectional BHF-FHS and prospective WOSCOPS. Finally, we undertook functional analysis of Y chromosome effects on monocyte and macrophage transcriptome in British men from the Cardiogenics Study.
Findings Of nine haplogroups identified, two (R1b1b2 and I) accounted for roughly 90% of the Y chromosome variants among British men. Carriers of haplogroup I had about a 50% higher age-adjusted risk of coronary artery disease than did men with other Y chromosome lineages in BHF-FHS (odds ratio 1·75, 95% CI 1·20—2·54, p=0·004), WOSCOPS (1·45, 1·08—1·95, p=0·012), and joint analysis of both populations (1·56, 1·24—1·97, p=0·0002). The association between haplogroup I and increased risk of coronary artery disease was independent of traditional cardiovascular and socioeconomic risk factors. Analysis of macrophage transcriptome in the Cardiogenics Study revealed that 19 molecular pathways showing strong differential expression between men with haplogroup I and other lineages of the Y chromosome were interconnected by common genes related to inflammation and immunity, and that some of them have a strong relevance to atherosclerosis.
Interpretation The human Y chromosome is associated with risk of coronary artery disease in men of European ancestry, possibly through interactions of immunity and inflammation.
Link
John Hawks on Neandertal similarity
In Which population in the 1000 Genomes Project samples has the most Neandertal similarity? John Hawks publishes some figures showing how different populations from the 1000 Genomes Project are more/less similar to the Vindija Neandertal sequence. My most recent thoughts on the topic on Neandertal admixture can be found here.
The most important findings are the following:
First, Tuscans appear to have more shared derived variants with Vindija than Brits do. I can think of two explanations for this finding:
- Higher genetic diversity in southern Europe compared to northern Europe; if these shared variants occurred at a low frequency in southern Europe vs. northern Europe to begin with, then genetic drift would have driven more of them to extinction in the north than in the south.
- Possibility that the Vindija sequence has modern human admixture from a population of early Southern Europeans that would have contributed more to the gene pool of Tuscans (who are geographically quite close to Croatia) than to Brits
Second, North Chinese appear to be more similar to Neandertals than South Chinese are.
- I am not entirely sure whether genetic drift would work in this case, since presumably CHB is representative of a larger Chinese population than Chinese settlers in Singapore.
- A different explanation is that south Chinese may possess more admixture from an Asian type of erectus-like population that predates the common ancestor of modern humans and Neandertals, who almost certainly lived in the western end of Eurafrasian region.
Third, the Luhya from Kenya appear to have less Neandertal similarity than the Yoruba from Nigeria. This, of course, makes very little sense if Neandertal similarity can only be attributed to Neandertal admixture, since there were no Neandertals in Africa at all. Moreover, while the Luhya live in Kenya, their ultimate origins are further west, since they are a Bantu group, although they show mixed West/East African affiliations.
There are two alternatives:
- Modern humans did originate in North-West Africa as the Y-chromosome phylogeny suggests, and East Africans possess some distinctive Palaeo-African ancestry from before the sapiens-Neandertal common ancestor.
- Back-migration from Eurasia that affected different populations to different degrees; it is my impression, for example, that the Yoruba are almost completely a Y-haplogroup E population, but if anyone has any comparative results at hand, feel free to comment.
February 08, 2012
Links between Native Americans and southern Altaians
AJHG doi:10.1016/j.ajhg.2011.12.014,
Mitochondrial DNA and Y Chromosome Variation Provides Evidence for a Recent Common Ancestry between Native Americans and Indigenous Altaians
Matthew C. Dulik et al.
The Altai region of southern Siberia has played a critical role in the peopling of northern Asia as an entry point into Siberia and a possible homeland for ancestral Native Americans. It has an old and rich history because humans have inhabited this area since the Paleolithic. Today, the Altai region is home to numerous Turkic-speaking ethnic groups, which have been divided into northern and southern clusters based on linguistic, cultural, and anthropological traits. To untangle Altaian genetic histories, we analyzed mtDNA and Y chromosome variation in northern and southern Altaian populations. All mtDNAs were assayed by PCR-RFLP analysis and control region sequencing, and the nonrecombining portion of the Y chromosome was scored for more than 100 biallelic markers and 17 Y-STRs. Based on these data, we noted differences in the origin and population history of Altaian ethnic groups, with northern Altaians appearing more like Yeniseian, Ugric, and Samoyedic speakers to the north, and southern Altaians having greater affinities to other Turkic speaking populations of southern Siberia and Central Asia. Moreover, high-resolution analysis of Y chromosome haplogroup Q has allowed us to reshape the phylogeny of this branch, making connections between populations of the New World and Old World more apparent and demonstrating that southern Altaians and Native Americans share a recent common ancestor. These results greatly enhance our understanding of the peopling of Siberia and the Americas.
Link
Mitochondrial DNA and Y Chromosome Variation Provides Evidence for a Recent Common Ancestry between Native Americans and Indigenous Altaians
Matthew C. Dulik et al.
The Altai region of southern Siberia has played a critical role in the peopling of northern Asia as an entry point into Siberia and a possible homeland for ancestral Native Americans. It has an old and rich history because humans have inhabited this area since the Paleolithic. Today, the Altai region is home to numerous Turkic-speaking ethnic groups, which have been divided into northern and southern clusters based on linguistic, cultural, and anthropological traits. To untangle Altaian genetic histories, we analyzed mtDNA and Y chromosome variation in northern and southern Altaian populations. All mtDNAs were assayed by PCR-RFLP analysis and control region sequencing, and the nonrecombining portion of the Y chromosome was scored for more than 100 biallelic markers and 17 Y-STRs. Based on these data, we noted differences in the origin and population history of Altaian ethnic groups, with northern Altaians appearing more like Yeniseian, Ugric, and Samoyedic speakers to the north, and southern Altaians having greater affinities to other Turkic speaking populations of southern Siberia and Central Asia. Moreover, high-resolution analysis of Y chromosome haplogroup Q has allowed us to reshape the phylogeny of this branch, making connections between populations of the New World and Old World more apparent and demonstrating that southern Altaians and Native Americans share a recent common ancestor. These results greatly enhance our understanding of the peopling of Siberia and the Americas.
Link
Big eye sockets compensate for low ambient light levels in humans
Biology Letters doi: 10.1098/rsbl.2011.0570
Latitudinal variation in light levels drives human visual system size
Eiluned Pearce and Robin Dunbar
Ambient light levels influence visual system size in birds and primates. Here, we argue that the same is true for humans. Light levels, in terms of both the amount of light hitting the Earth's surface and day length, decrease with increasing latitude. We demonstrate a significant positive relationship between absolute latitude and human orbital volume, an index of eyeball size. Owing to tight scaling between visual system components, this will translate into enlarged visual cortices at higher latitudes. We also show that visual acuity measured under full-daylight conditions is constant across latitudes, indicating that selection for larger visual systems has mitigated the effect of reduced ambient light levels. This provides, to our knowledge, the first support that light levels drive intraspecific variation in visual system size in the human population.
Link
Latitudinal variation in light levels drives human visual system size
Eiluned Pearce and Robin Dunbar
Ambient light levels influence visual system size in birds and primates. Here, we argue that the same is true for humans. Light levels, in terms of both the amount of light hitting the Earth's surface and day length, decrease with increasing latitude. We demonstrate a significant positive relationship between absolute latitude and human orbital volume, an index of eyeball size. Owing to tight scaling between visual system components, this will translate into enlarged visual cortices at higher latitudes. We also show that visual acuity measured under full-daylight conditions is constant across latitudes, indicating that selection for larger visual systems has mitigated the effect of reduced ambient light levels. This provides, to our knowledge, the first support that light levels drive intraspecific variation in visual system size in the human population.
Link
42,000 year old art from Andalusia
'The oldest work of art ever': 42,000-year-old paintings of seals found in Spanish cave
Not sure whether this has been (or will be) published elsewhere, or what is the evidence, exactly, that this art was made by Neandertals. If the date is correct, it is certainly a spooky coincidence with the appearance of the first modern humans in Europe.The world's oldest works of art have been found in a cave on Spain's Costa del Sol, scientists believe. Six paintings of seals are at least 42,000 years old and are the only known artistic images created by Neanderthal man, experts claim. Professor Jose Luis Sanchidrian, from the University of Cordoba, described the discovery as 'an academic bombshell', as all previous art work has been attributed to Homo sapiens.
The paintings were found in the Nerja Caves, 35 miles east of Malaga in the southern region of Andalusia.Spanish scientists sent organic residue found next to the paintings to Miami, where they were dated at being between 43,500 and 42,300 years old.
UPDATE (Feb 23): A more detailed story from the New Scientist.
February 07, 2012
High coverage Denisovan genome
With 30-fold coverage from here.
Let this be a lesson to all those who have been sitting on important data (*cough* Otzi's genome) for more than a year.
Approximately 30-fold coverage of the genome was generated using the Illumina GAIIx sequencing platform.
We provide here the additional high-coverage sequence data which augments the previously published sequence data.
This data is freely available without passwords.
The data represents sequence data generated on the Illumina GAIIx platform and constitutes an average 30-fold coverage of the Denisovan genome. Focussing on the "mappable genome" - the 1.86GB of unique sequence with clear human-chimpanzee orthology - we see 99.97% of bases covered at least 3x and 99.51% covered at least 10x.
Contamination with modern human DNA, estimated from both mitochondrial and nuclear genomes, is less than 1%and:
The Denisova Genome Consortium have chosen to release the raw sequence data and alignments for the additional sequence generated for the Denisova phalanx (Reich et al. Science. 2010) in the expectation that these data will be a valuable resource for the research community.
In keeping with the Ft. Lauderdale principles, the data are available for use, but users are expected to allow the data producers to make the first presentations and to publish the first paper containing genome-wide analyses of the data. Researchers who use small amounts of the data (eg: for single locus analyses) are not required to request permission. Researchers who have queries about whether they may present or submit Denisova genome data for publication may contact Svante Paabo.This is great, since it is unusual for scientists to publish data before they publish the paper describing the data. This gives scientists and non-scientists alike the opportunity to start working with the data, and to begin the development of new ideas.
Let this be a lesson to all those who have been sitting on important data (*cough* Otzi's genome) for more than a year.
February 06, 2012
No bottleneck leading to Homo sapiens in Africa
This is continuation of previous work by many of the same authors. They consider two alternative models of human origins:
- A bottleneck that left only a small number of humans in Africa during the penultimate glacial age (130-190kya)
- Fragmentation of the range occupied by humans during that time, which led to a number of different groups surviving, but becoming separated by uninhabitable areas
As I've mentioned before on the blog, the authors write that:
The choice of mutation rate can impact the results and because there is a 2-3 fold di erence between the mutation rate estimated from mother-father-child trios (or quartet) ( ~10^-8=bp=generation; The 1000 Genomes Project Consortium 2010; Roach et al. 2010) and the mutation rate derived from human-chimp comparisons ( ~2.5x10^-8=bp=generation; Nachman and Crowell 2000; Fagundes et al. 2007; Gutenkunst et al. 2009; Laval et al. 2010), we considered both estimates of the mutation rate in our analyses.
There is, in fact, an interplay between three variables: time, mutation rate, and effective population size. If one fixes time and chooses a mutation rate, then they can arrive at an estimate of effective population size. This is, in fact, what the authors do, and they conclude that:
Our fi ndings are consistent with this hypothesis, but, depending on the mutation rate, we find either an eff ective population size of NA = 12,000 (95%C.I. = 9,000-15; 500 when averaging over all three demographic models) using the mutation rate calibrated with the human-chimp divergence or an e ffective population size of NA = 32,500 individuals (95%C:I: = 27,500-34,500) using the mutation rate given by whole-genome trio analysis (The 1000 Genomes Project Consortium 2010) (supplementary fi gure 4 and table 6, Supplementary Material online).I believe that in the coming years our understanding of deep human prehistory will be aided by two developments:
...
Relating the estimated eff ective population size to the census population size during the Pleistocene is a di cult task because there are many factors a ecting the e ective population size (Charlesworth 2009). Nevertheless, based on published estimates of the ratio between e ffective and census population size, a comprehensive value on the order of 10% has been found by Frankham (1995). This 10% rule roughly predicts that 120,000-325,000 individuals (depending on the assumed mutation rate) lived in Sub-Saharan Africa some 130 kya. Assuming that the range of humans extends over all the 24 millions km2 of Sub-Saharan Africa, the density of humans at that time would have been extremely low between 0.5 and 1.4 individual per 100 km2, which is even lower than the lowest recorded hunter gatherer density of 2 individuals per 100 km2 reported for the !Kung (Kelly 1995) and the density of 3 individuals per 100 km2 estimated for Middle Paleolithic people (Hassan 1981). However, this discrepancy disappears if humans were restricted to an area some 3-6 times smaller than the entire Sub-Saharan Africa.
- First, we will be able to estimate the autosomal mutation rate very robustly from a large number of families, so we will no longer have to calibrate it using paleontological data points such as periods of glaciation or the human-chimp divergence
- Second, we will be able to obtain ancient DNA from Pleistocene era modern humans, and hence incorporate real data points into models of human evolution
- A plethora of modern humans in a "Green Sahara" prior to the most recent Ice Age, which expanded into the Near East and gave us Skhul/Qafzeh/Jebel Faya/Nubian Complex.
- A plethora of modern humans in a "Green Arabia/Gulf Oasis" prior to 70kya that experienced a population crash due to desertification post-70kya and led to an exodus of modern humans into the rest of Eurasia, and back-to-Africa
A Green Sahara would fit the bill for a geographically limited area within Africa that would support a large population at comparable densities to modern hunter-gatherer populations of Africa. Ultimately, the puzzle of human origins can only be solved by a combination of genetics, archaeology, and palaeoanthropology.
Mol Biol Evol (2012)doi: 10.1093/molbev/mss061
Resequencing data provide no evidence for a human bottleneck in Africa during the penultimate glacial period
Per Sjödin, Agnès E Sjöstrand, Mattias Jakobsson and Michael G B Blum
Based on the accumulation of genetic, climatic and fossil evidence, a central theory in paleoanthropology stipulates that a demographic bottleneck coincides with the origin of our species Homo Sapiens. This theory proposes that anatomically modern humans – which were only present in Africa at the time – experienced a drastic bottleneck during the penultimate glacial age (130-190 kya) when a cold and dry climate prevailed. Two scenarios have been proposed to describe the bottleneck, which involve either a fragmentation of the range occupied by humans or the survival of one small group of humans. Here, we analyze DNA sequence data from 61 nuclear loci sequenced in three African populations using Approximate Bayesian Computation and numerical simulations. In contrast to the bottleneck theory, we show that a simple model without any bottleneck during the penultimate ice age has the greatest statistical support compared to bottleneck models. Although the proposed bottleneck is ancient, occurring at least 130 kya, we can discard the possibility that it did not leave detectable footprints in the DNA sequence data except if the bottleneck involves a less than a 3 fold reduction in population size. Finally, we confirm that a simple model without a bottleneck is able to reproduce the main features of the observed patterns of genetic variation. We conclude that models of Pleistocene refugium for modern human origins now require substantial revision.
February 03, 2012
Y-chromosome admixture in self-identified Australian Aboriginals
Forensic Sci Int Genet. 2012 Jan 30. [Epub ahead of print]
An investigation of admixture in an Australian Aboriginal Y-chromosome STR database.
Taylor D, Nagle N, Ballantyne KN, van Oorschot RA, Wilcox S, Henry J, Turakulov R, Mitchell RJ.
Abstract
Y-chromosome specific STR profiling is increasingly used in forensic casework. However, the strong geographic clustering of Y haplogroups can lead to large differences in Y-STR haplotype frequencies between different ethnicities, which may have an impact on database composition in admixed populations. Aboriginal people have inhabited Australia for over 40,000 years and until ∼300 years ago they lived in almost complete isolation. Since the late 18th century Australia has experienced massive immigration, mainly from Europe, although in recent times from more widespread origins. This colonisation resulted in highly asymmetrical admixture between the immigrants and the indigenes. A State jurisdiction within Australia has created an Aboriginal Y-STR database in which assignment of ethnicity was by self-declaration. This criterion means that some males who identify culturally as members of a particular ethnic group may have a Y haplogroup characteristic of another ethnic group, as a result of admixture in their paternal line. As this may be frequent in Australia, an examination of the extent of genetic admixture within the database was performed. A Y haplogroup predictor program was first used to identify Y haplotypes that could be assigned to a European haplogroup. Of the 757 males (589 unique haplotypes), 445 (58.8%) were identified as European (354 haplotypes). The 312 non-assigned males (235 haplotypes) were then typed, in a hierarchical fashion, with a Y-SNP panel that detected the major Y haplogroups, C-S, as well as the Aboriginal subgroup of C, C4. Among these 96 males were found to have non-Aboriginal haplogroups. In total, ∼70% of Y chromosomes in the Aboriginal database could be classed as non-indigenous, with only 169 (129 unique haplotypes) or 22% of the total being associated with haplogroups denoting Aboriginal ancestry, C4 and K* or more correctly K(xL,M,N,O,P,Q,R,S). The relative frequencies of these indigenous haplogroups in South Australia (S.A.) were significantly different to those seen in samples from the Northern Territory and Western Australia. In S.A., K* (∼60%) has a much higher frequency than C4 (∼40%), and the subgroup of C4, C4(DYS390.1del), comprised only 17%. Clearly admixture in the paternal line is at high levels among males who identify themselves as Australian Aboriginals and this knowledge may have implications for the compilation and use of Y-STR databases in frequency estimates.
Link
An investigation of admixture in an Australian Aboriginal Y-chromosome STR database.
Taylor D, Nagle N, Ballantyne KN, van Oorschot RA, Wilcox S, Henry J, Turakulov R, Mitchell RJ.
Abstract
Y-chromosome specific STR profiling is increasingly used in forensic casework. However, the strong geographic clustering of Y haplogroups can lead to large differences in Y-STR haplotype frequencies between different ethnicities, which may have an impact on database composition in admixed populations. Aboriginal people have inhabited Australia for over 40,000 years and until ∼300 years ago they lived in almost complete isolation. Since the late 18th century Australia has experienced massive immigration, mainly from Europe, although in recent times from more widespread origins. This colonisation resulted in highly asymmetrical admixture between the immigrants and the indigenes. A State jurisdiction within Australia has created an Aboriginal Y-STR database in which assignment of ethnicity was by self-declaration. This criterion means that some males who identify culturally as members of a particular ethnic group may have a Y haplogroup characteristic of another ethnic group, as a result of admixture in their paternal line. As this may be frequent in Australia, an examination of the extent of genetic admixture within the database was performed. A Y haplogroup predictor program was first used to identify Y haplotypes that could be assigned to a European haplogroup. Of the 757 males (589 unique haplotypes), 445 (58.8%) were identified as European (354 haplotypes). The 312 non-assigned males (235 haplotypes) were then typed, in a hierarchical fashion, with a Y-SNP panel that detected the major Y haplogroups, C-S, as well as the Aboriginal subgroup of C, C4. Among these 96 males were found to have non-Aboriginal haplogroups. In total, ∼70% of Y chromosomes in the Aboriginal database could be classed as non-indigenous, with only 169 (129 unique haplotypes) or 22% of the total being associated with haplogroups denoting Aboriginal ancestry, C4 and K* or more correctly K(xL,M,N,O,P,Q,R,S). The relative frequencies of these indigenous haplogroups in South Australia (S.A.) were significantly different to those seen in samples from the Northern Territory and Western Australia. In S.A., K* (∼60%) has a much higher frequency than C4 (∼40%), and the subgroup of C4, C4(DYS390.1del), comprised only 17%. Clearly admixture in the paternal line is at high levels among males who identify themselves as Australian Aboriginals and this knowledge may have implications for the compilation and use of Y-STR databases in frequency estimates.
Link
February 01, 2012
A timeline of human prehistory
I will be using the following equation to estimate some times in human prehistory:
Time t can be estimated if one has an estimate of the effective population size Ne, and of Fst between a pair of ancestral populations. We also need some estimate of the generation length to convert times into years.
I take the long-term human generation length to be 23 years (Hawks et al. 2000)
I will use the Fst values recently inferred by myself using ADMIXTURE on a large dataset of Old World variation; I note that this dataset did not include African hunter gatherers.
I use the estimates of Ne by Li et al. My procedure is as follows: I note which population is modal for each of the 12 ancestral components, and note its Ne. I take the average of the two populations for which there is an Fst value.
For example, the Atlantic_Med component is modal in the HGDP French_Basque population (Ne=6,137), and the Sub_Saharan in the HGDP Yoruba population (Ne=9,960). The average Ne=8,049. The Fst betwen these two components is 0.185. Hence, the time of divergence is:
23*log(1-0.185)/log(1-1/(2*8049)) = 75.7ky
Below are the pairwise divergence times of the maximally differentiated components: Atlantic_Med, Southeast_Asian, Sub_Saharan; within the HGDP, these are modal in French_Basque, Dai, and Yoruba
Atlantic_Med vs. Sub_Saharan = 75.7ky
Atlantic_Med vs. Southeast_Asian = 43.0ky
Sub_Saharan vs. Southeast_Asian = 86.4ky
These dates appear plausible. An important calibration point is the appearance of the first modern humans in Europe which has been archaeologically placed at precisely the time given above for the Atlantic_Med vs. Southeast_Asian divergence.
We can probably assume that the differentiation between Eurasians and Sub-Saharan Africans is best estimated by the Sub_Saharan vs. Southeast_Asian comparison, since Southeast_Asians would have extremely limited opportunity to experience subsequent gene flow with Africans in either direction. The time of 86.4ky is older than the 70ky estimated for mtDNA haplogroup L3 which almost certainly marks the major human population expansion. The difference can be probably attributed to the absorption of different populations in Africa and Asia (archaic humans or pre-L3 expansion modern humans).
Of particular interest to me is an evaluation of my womb of nations theory, with respect to the origin of West Eurasians. The Caucasus component exhibits Fst distances between 0.033 and 0.052 with the components I have labeled "the Six". Within the HGDP, the following populations are modal for them:
Caucasus: Adygei; Atlantic_Med: French_Basque; North_European: Orcadian; Southwest_Asian: Bedouin; Northwest_African: Mozabite; Gedrosia: Brahui
Divergence times are:
Caucasus vs. Atlantic_Med = 9.9ky
Caucasus vs. North_European = 11.1ky
Caucasus vs. Southwest_Asian = 16.8ky
Caucasus vs. Northwest_African = 16.2ky
Caucasus vs. Gedrosia = 13.6ky
There is reason to think that the last 3 ages are inflated: the Northwest_African and Gedrosia component appear divergent in a Sub_Saharan and South_Asian direction respectively, so they probably represent stabilized mixtures of a West Eurasian with a substrate element. Moreover, the effective population size used for the Bedouin is quite higher than for any other West Eurasian population, presumably because of the African admixture in that population; that high effective size is, hence, not reflective of the effective size of the Southwest_Asian ancestral component.
All in all, these results are consistent with a mainly Neolithic origin of West Eurasian populations, and incompatible with their pre-LGM differentiation. As I have argued in the womb of nations, the expanding Neolithic population probably absorbed -to a limited extent- aborigines from outside the core area of the Near East, however, the bulk of the population appears to share a fairly recent common ancestry.
Time t can be estimated if one has an estimate of the effective population size Ne, and of Fst between a pair of ancestral populations. We also need some estimate of the generation length to convert times into years.
I take the long-term human generation length to be 23 years (Hawks et al. 2000)
I will use the Fst values recently inferred by myself using ADMIXTURE on a large dataset of Old World variation; I note that this dataset did not include African hunter gatherers.
I use the estimates of Ne by Li et al. My procedure is as follows: I note which population is modal for each of the 12 ancestral components, and note its Ne. I take the average of the two populations for which there is an Fst value.
For example, the Atlantic_Med component is modal in the HGDP French_Basque population (Ne=6,137), and the Sub_Saharan in the HGDP Yoruba population (Ne=9,960). The average Ne=8,049. The Fst betwen these two components is 0.185. Hence, the time of divergence is:
23*log(1-0.185)/log(1-1/(2*8049)) = 75.7ky
Below are the pairwise divergence times of the maximally differentiated components: Atlantic_Med, Southeast_Asian, Sub_Saharan; within the HGDP, these are modal in French_Basque, Dai, and Yoruba
Atlantic_Med vs. Sub_Saharan = 75.7ky
Atlantic_Med vs. Southeast_Asian = 43.0ky
Sub_Saharan vs. Southeast_Asian = 86.4ky
These dates appear plausible. An important calibration point is the appearance of the first modern humans in Europe which has been archaeologically placed at precisely the time given above for the Atlantic_Med vs. Southeast_Asian divergence.
We can probably assume that the differentiation between Eurasians and Sub-Saharan Africans is best estimated by the Sub_Saharan vs. Southeast_Asian comparison, since Southeast_Asians would have extremely limited opportunity to experience subsequent gene flow with Africans in either direction. The time of 86.4ky is older than the 70ky estimated for mtDNA haplogroup L3 which almost certainly marks the major human population expansion. The difference can be probably attributed to the absorption of different populations in Africa and Asia (archaic humans or pre-L3 expansion modern humans).
Of particular interest to me is an evaluation of my womb of nations theory, with respect to the origin of West Eurasians. The Caucasus component exhibits Fst distances between 0.033 and 0.052 with the components I have labeled "the Six". Within the HGDP, the following populations are modal for them:
Caucasus: Adygei; Atlantic_Med: French_Basque; North_European: Orcadian; Southwest_Asian: Bedouin; Northwest_African: Mozabite; Gedrosia: Brahui
Divergence times are:
Caucasus vs. Atlantic_Med = 9.9ky
Caucasus vs. North_European = 11.1ky
Caucasus vs. Southwest_Asian = 16.8ky
Caucasus vs. Northwest_African = 16.2ky
Caucasus vs. Gedrosia = 13.6ky
There is reason to think that the last 3 ages are inflated: the Northwest_African and Gedrosia component appear divergent in a Sub_Saharan and South_Asian direction respectively, so they probably represent stabilized mixtures of a West Eurasian with a substrate element. Moreover, the effective population size used for the Bedouin is quite higher than for any other West Eurasian population, presumably because of the African admixture in that population; that high effective size is, hence, not reflective of the effective size of the Southwest_Asian ancestral component.
All in all, these results are consistent with a mainly Neolithic origin of West Eurasian populations, and incompatible with their pre-LGM differentiation. As I have argued in the womb of nations, the expanding Neolithic population probably absorbed -to a limited extent- aborigines from outside the core area of the Near East, however, the bulk of the population appears to share a fairly recent common ancestry.


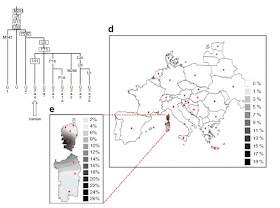
















.png)






