The data can be found here. Description of region codes:
Cornwall, Devon and Pembrokeshire were pooled to represent the South/West (SW) and the area that could be considered the closest surrogate to the Ancient British. Kent, Norfolk and Lincolnshire were pooled to represent the East (E) and the area most directly influenced by the Anglo-Saxon invasions. Cumbria, Yorkshire and the North East were pooled broadly to represent the North of England (N); Oxfordshire and the Forest of Dean were combined to represent the Central region of England (CN); and Orkney was kept separate from the others, largely because of the known substantial Norse Viking influence in Orkney.
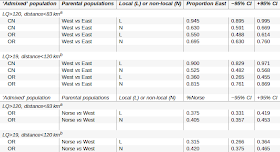
Of interest are the NRY haplogroup data:
The SW seems to have the lowest R1a1 frequency (1.3%). This is in agreement with the Dodecad v3 results for Cornwall (1.8% East European). The E seems to have an intermediate frequency (3.5%), again in agreement with Dodecad v3 for Kent (3.7%). Orkney has the highest R1a1 frequency (34.2%) and also the highest East European component in Dodecad v3 (11%). So, it does seem that R1a1 frequency tracks an eastern population element in the British isles.
R1xR1a1 has its lowest values in E and OR; this is probably consistent with the idea that Germanic invaders from the east possessed less of this haplogroup than the pre-Germanic population.
The minor alleles in MC1R are associated with red hair, and it seems that this is maximized in Orkney.
The admixture estimates are also quite interesting, showing a dependence on the use of local (L) vs. non-local (N) surnames; the results do suggest non-trivial shifts in genetic composition since the adoption of surnames.
Also of interest: A British approach to sampling
European Journal of Human Genetics advance online publication 10 August 2011; doi: 10.1038/ejhg.2011.127
People of the British Isles: preliminary analysis of genotypes and surnames in a UK-control population
Bruce Winney et al.
There is a great deal of interest in a fine-scale population structure in the UK, both as a signature of historical immigration events and because of the effect population structure may have on disease association studies. Although population structure appears to have a minor impact on the current generation of genome-wide association studies, it is likely to have a significant part in the next generation of studies designed to search for rare variants. A powerful way of detecting such structure is to control and document carefully the provenance of the samples involved. In this study, we describe the collection of a cohort of rural UK samples (The People of the British Isles), aimed at providing a well-characterised UK-control population that can be used as a resource by the research community, as well as providing a fine-scale genetic information on the British population. So far, some 4000 samples have been collected, the majority of which fit the criteria of coming from a rural area and having all four grandparents from approximately the same area. Analysis of the first 3865 samples that have been geocoded indicates that 75% have a mean distance between grandparental places of birth of 37.3 km, and that about 70% of grandparental places of birth can be classed as rural. Preliminary genotyping of 1057 samples demonstrates the value of these samples for investigating a fine-scale population structure within the UK, and shows how this can be enhanced by the use of surnames.
Link



Hmm
ReplyDeleteWhy is the South West considered the proxy for the most ancient Britons? North has the highest haplogroup I (beleived to be the oldest haplogroup). By a substantial margin.
At first glance,one of the more important findings seems to be that hap I is not related to east vs west in England. The only group with unusual levels of I are the northern English.
ReplyDeleteIf we go by history maybe the Anglo Saxons (immigration varies from east to west) didnt have a big impact on I but the Vikings (mostly in the north) did.
Of course there might be more to it if we say the break down of I.
Did the researchers cover Northern Ireland? It would be interesting to see the genetic differences between the Catholics and the Protestants. My hunch is that the Catholics have higher levels of R1b.
ReplyDeleteAnother paper of supremely useless nature, as it did not distinguish between Hg I subclades.
ReplyDeleteIt is well-understood that I1 distinguishes Scandinavians and I2 the more Western and Southwestern / Atlantic Seaboard early Europeans.
But no, the paper just lists generic I. That is why the "original Britons" population (expected to be much I2) and the "expected Viking and Anglo-Saxon" (expected to be much I1) populations have roughly the same numbers...
Would anyone read a paper that had generic R1 stats? (Not distinguishing between R1a and R1b)? No, it would be senseless. Oh that's right, no one would ever publish such a paper because researchers are very R1-centric. Wish they'd do the same for I.
I was going to say there were more authors than samples, but that would be a slight exaggeration.
ReplyDeleteAbout 440 samples to base these results on?
North has the highest haplogroup I (beleived to be the oldest haplogroup).
ReplyDeleteWhoever said that Haplogroup I is the oldest one?
A conference presentation covering what is in this paper as well as an explanation of regional differences included the finding that Devon and Cornwall are genetically distinct from the rest of England, along with the presenter's belief that the people of western Britain are closest to the first post-Ice Age colonizers. That is why Devon and Cornwall are a proxy for the most ancient Britons. I don't know why Pembrokeshire was included as part of the proxy in the published paper. Presumably that will be made clear in the next paper.
ReplyDeleteThe results for Orkney are interesting. 34% R1a, which is 6 times higher than Central England and 15 times higher than Northern England! And the 8% Orkney level of I is about half that of anywhere on the mainland. That would imply that the Vikings who settled there had high R1a and low I. Yet Norway has higher levels of I than R1a. This perhaps implies that Norway was not uniform in the Viking period. It also suggests that high levels of Germanic migration need not necessarily leave high levels of I -- it can actually leave much lower levels of I!
ReplyDeleteIf the I was coming from th Vikings it would be in the Orkneys and it isnt.
ReplyDeletePlus in this paper, amazingly, they did NOT distinguish between R1a and R1b. Which I would have thought of as a minimum discrimination. Surely the raw data is bettter than this?
Still I am surprised they are seeing such big differences. I increasingly think that Y-haplogroups are a very poor method of understanding populations. I think they can be massively amplified out of proportion to the real population differences.
"Why is the South West considered the proxy for the most ancient Britons? North has the highest haplogroup I (beleived to be the oldest haplogroup). By a substantial margin".
ReplyDeletePerhaps, as Average Joe said, I is not the oldest haplogroup in the British Isles.
"My hunch is that the Catholics have higher levels of R1b".
It is certainly quite likely to be more common in the west Off Ireland as a study years ago showed that nearly all men in the west with Irish surnames had that haplogroup:
http://homepage.eircom.net/~oflannery/YHILL2000.pdf
"Devon and Cornwall are genetically distinct from the rest of England, along with the presenter's belief that the people of western Britain are closest to the first post-Ice Age colonizers".
Makes sense to me. Most immigrants would have arrived on the east coast first.
"they did NOT distinguish between R1a and R1b".
They did distinguish between R1a1 and all other Rs, presumably all R1b.
"I increasingly think that Y-haplogroups are a very poor method of understanding populations. I think they can be massively amplified out of proportion to the real population differences".
True. They only tell us about male movement, and may not tell us too much about ancient ones at that.
"If the I was coming from th Vikings it would be in the Orkneys and it isnt."
ReplyDeleteWell, my logic was that the pre Viking English were nearly the same and that the Viking added hap I to the northern English resulting in the modern differences.
We don't know what the pre Viking orkneys had and the Viking could have added hap I if you assume a lower number than today.
"I increasingly think that Y-haplogroups are a very poor method of understanding populations."
ReplyDeleteFor most questions... Ya
"they did NOT distinguish between R1a and R1b. Which I would have thought of as a minimum discrimination."
ReplyDeleteBut they list R1a1 seperate.
With such a small sample of 400 males how can one infer that either I1 or any of the R's were the most ancient populations in Britain. That being said, it is known that Britain was connected to Europe over 8000 years ago and prior to that its unknown how many various populations once roamed the British isles. Likewise those populations may or may not have survived in the isle to today.
ReplyDeleteIf they used high-resolution studies combined with known census and historical records...it could tell you plenty.
ReplyDeleteDevon and Cornwall are genetically distinct from the rest of England, along with the presenter's belief that the people of western Britain are closest to the first post-Ice Age colonizers".
ReplyDeleteThen they really should have included Ireland, the ultimate West.
But as understand it folk moved along the coast mostly from the south. So inland, remote islands, north west Ireland, Northwest England and Western Scotland should be the last to receive folk. Nothing in southern england can really be considered remote. There counties are no more than two days ride from London.
Cornwall is particularly interesting because it managed to retain its language (and presumably some of its culture) for the longest. But this is associated with French Brittany (or vice versa). So clearly there was a lot of population congress between the two in relatively recent times. This does not equate to oldest.
The F without I, J2 or R1 is probably a measure of G2 which has been so interesting lately.
ReplyDeleteLater immigrant haplogroups can be expected to be higher in the East (as this F is). Which makes the result for the F in the central region more interesting, as it is low in the other immigrants E and J2. I wonder what it means.
Then they really should have included Ireland, the ultimate West
ReplyDeleteBut the research is on British DNA, not Irish genetics.
"But the research is on British DNA, not Irish genetics."
ReplyDeleteI think the point is that that is a political rather than a genetic distinction.
Annie makes an important point - western Britain may be relatively remote to Geremany and that coast, but that doesn't make it remote. It is close to France and in fact in Roman times Ireland the most active trade routes put it closer to Rome and the Mediterranean in general than Kent/London/East Anglia were.
And this was not just a function of Roman rule. The legend is that the Irish language came to Ireland from northern Spain, with no mention of Roamn ruel in Spain at the time.
"If the I was coming from th Vikings it would be in the Orkneys and it isnt."
ReplyDeleteI don't think the nortern grouping makes sense. From the OP
"Cumbria, Yorkshire and the North East were pooled broadly to represent the North of England (N)"
Cumbria (note the name - Wales is Cymru in Welsh) is similar to Cornwall. It was one of the last Romano-British kingdoms (Rheged) to fall to the Anglo-Saxons and is relatively remote so the "I" values may be coming from that.
I'd have thought Cumbria would make more sense in the Southwest group.