Nature 451, 998-1003 (21 February 2008) | doi:10.1038/nature06742; Received 2 December 2007; Accepted 29 January 2008
Genotype, haplotype and copy-number variation in worldwide human populations
Mattias Jakobsson1,2,14, Sonja W. Scholz4,5,14, Paul Scheet1,3,14, J. Raphael Gibbs4,5, Jenna M. VanLiere1, Hon-Chung Fung4,6, Zachary A. Szpiech1, James H. Degnan1,2, Kai Wang7, Rita Guerreiro4,8, Jose M. Bras4,8, Jennifer C. Schymick4,9, Dena G. Hernandez4, Bryan J. Traynor4,10, Javier Simon-Sanchez4,11, Mar Matarin4, Angela Britton4, Joyce van de Leemput4,5, Ian Rafferty4, Maja Bucan7, Howard M. Cann12, John A. Hardy5, Noah A. Rosenberg1,2,3 & Andrew B. Singleton4,13
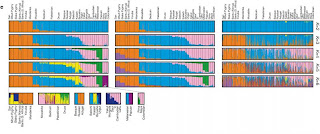
Genome-wide patterns of variation across individuals provide a powerful source of data for uncovering the history of migration, range expansion, and adaptation of the human species. However, high-resolution surveys of variation in genotype, haplotype and copy number have generally focused on a small number of population groups1, 2, 3. Here we report the analysis of high-quality genotypes at 525,910 single-nucleotide polymorphisms (SNPs) and 396 copy-number-variable loci in a worldwide sample of 29 populations. Analysis of SNP genotypes yields strongly supported fine-scale inferences about population structure. Increasing linkage disequilibrium is observed with increasing geographic distance from Africa, as expected under a serial founder effect for the out-of-Africa spread of human populations. New approaches for haplotype analysis produce inferences about population structure that complement results based on unphased SNPs. Despite a difference from SNPs in the frequency spectrum of the copy-number variants (CNVs) detected—including a comparatively large number of CNVs in previously unexamined populations from Oceania and the Americas—the global distribution of CNVs largely accords with population structure analyses for SNP data sets of similar size. Our results produce new inferences about inter-population variation, support the utility of CNVs in human population-genetic research, and serve as a genomic resource for human-genetic studies in diverse worldwide populations.
Link

No comments:
Post a Comment
Stay on topic. Be polite. Use facts and arguments. Be Brief. Do not post back to back comments in the same thread, unless you absolutely have to. Don't quote excessively. Google before you ask.