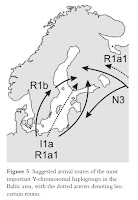
From the paper on Y-haplogroup I1a:
Haplogroup I1a is suggested to have its origins in the Iberian refugium, from where it spread northward and now has its highest frequencies in Northern Europe (Rootsi et al. 2004). The haplotype matches to Germany and Poland imply that I1a has arrived to the Nordic countries from the Southern Baltic Sea region, which is historically plausible. The coalescense age of the haplogroup is about 5000 years lower than the age of the earliest archaeological findings from the Northern Baltic Sea region, which suggests a Neolithic arrival. There are two possible migration routes from Central Europe to the Northern Baltic Sea region: an exclusive western route via Sweden, an eastern route via the Baltic states, or via both to Eastern Finland and Karelia (Fig. 5). The surprisingly high diversities of I1a among the eastern Finnish and Baltic populations, and the lack of association between the Western Finns and the Swedes in SAMOVA analysis suggest that I1a has been involved in bifurcating migrations both via Sweden and the Baltic states, and that the presence of the haplogroup in Finland and Karelia is not merely due to Swedish influence. The low frequency of I1a among the Baltic populations may be due to later effects of genetic drift or replacement.
I am personally doubtful of the Iberian origin of Y-haplogroup I1a. Its presence in Iberia and France, as well as its high diversity there may be the result of migration from northern Europe, a genetic trace of the Germanic Volkerwanderung. One certainly needs to consider this effect. There are probably a couple of papers just waiting to be written by looking at Y-chromosomes of Germanic descendants in Southwestern Europe by looking at either surnames or early cemetaries.
With regard to Y-haplogroup N3:
The frequency distribution and age of haplogroup N3 in our study sample was consistent with the earlier studies (Lahermo et al. 1999, Zerjal et al. 2001, Tambets et al. 2004, Karlsson et al. 2006, Rootsi et al. 2007). According to the YHRD database, the haplotypes most common in Finland and Karelia were relatively unique, which is not unexpected, since data from most Eurasian populations where N3 is common is not publicly available. It seems evident that the Finns and Karelians share a history regarding haplogroup N3. In the database comparisons, we also observed that N3 may mark a westward diffusion in the north from Finland to Sweden and in the south from the Baltic countries to Poland and Germany.The researchers also reiterated the previous idea of a dual origin of N3, as shown in the figure, with different clades being represented in Finland and the Baltic states, with Estonia being intermediate between the two.
With regard to Y-haplogroup R1a1:
It is plausible that both R1a1 and I1a were carried to the Baltic Sea region via the same Neolithic migrations from Germany/Poland. The higher coalescence age and the starlike network structure of R1a1 are consistent with the probable higher diversity and frequency of R1a1 in the original source population(s), a consequence of the wider geographical distribution of the haplogroup. It is an important observation that in the Baltic Sea region R1a1 is mainly associated to Central European rather than eastern or Russian influence. However, haplotype frequency comparisons (Derenko et al. 2006, Willuweit & Roewer 2007) give some indication of Russian gene flow as a partial source of R1a1 in Karelia, which would be plausible given the long period of admixture with Slavs (Fig. 5). However, the Y-chromosomal diversity in Karelia has been heavily affected by drift and founder effects. Another haplogroup with eastern affinity is I1b (Rootsi et al. 2004), whose presence in Karelia and the Baltic states is probably a sign of Russian gene flow.
This is an important discovery, and a great first step in uncovering the structure within this widespread haplogroup. Even though R1a1 Y-chromosomes were studied in a lot of populations, including e.g., the Balkans, India, the Altai, unfortunately we know next to nothing about its phylogenetic substructure. Without such knowledge, R1a1 spread has been variously interpreted as a signal of postglacial colonization, Kurgan expansions, or the spread of Slavic languages.
Finally, the genetic legacy of the Saami is visible in mtDNA:
The eastern elements in the mtDNA variation of the Baltic Sea region are intertwined with the Saami influence. Recent studies of the mtDNA variation among the Saami show a link to the Volga-Ural region (Tambets et al. 2004, Ingman & Gyllensten 2006), which is now shown to exist also among the Karelians and, to a lesser degree, among the other populations from the Baltic Sea region as well. Additionally, the presence of U4 in the Eastern Baltic Sea populations may represent eastern influence, since it is typical for the Volga-Ural region (Bermisheva et al. 2002). The high diversity of this haplogroup in the Baltic region, observable in the haplotype network, suggests a complex history, and rules genetic drift out as a cause of the high frequency. All in all, these mtDNA haplogroups may be maternal reflections of the eastern influence that can be most clearly observed in the Y-chromosomal haplogroup N3.
Annals of Human Genetics doi:10.1111/j.1469-1809.2007.00429.x
Migration Waves to the Baltic Sea Region
T. Lappalainen et al.
In this study, the population history of the Baltic Sea region, known to be affected by a variety of migrations and genetic barriers, was analyzed using both mitochondrial DNA and Y-chromosomal data. Over 1200 samples from Finland, Sweden, Karelia, Estonia, Setoland, Latvia and Lithuania were genotyped for 18 Y-chromosomal biallelic polymorphisms and 9 STRs, in addition to analyzing 17 coding region polymorphisms and the HVS1 region from the mtDNA. It was shown that the populations surrounding the Baltic Sea are genetically similar, which suggests that it has been an important route not only for cultural transmission but also for population migration. However, many of the migrations affecting the area from Central Europe, the Volga-Ural region and from Slavic populations have had a quantitatively different impact on the populations, and, furthermore, the effects of genetic drift have increased the differences between populations especially in the north. The possible explanations for the high frequencies of several haplogroups with an origin in the Iberian refugia (H1, U5b, I1a) are also discussed.
Link

No comments:
Post a Comment
Stay on topic. Be polite. Use facts and arguments. Be Brief. Do not post back to back comments in the same thread, unless you absolutely have to. Don't quote excessively. Google before you ask.