A very exciting new paper has just appeared in AJHG. The authors have used the 10,000+ SNP array by Affymetrix to study the individuals from the Y chromosome consortium panel. Note that these individuals were originally collected to study Y chromosomal variation, but in this case, they were studied because they represent a globally diverse set of people, and their genomic variation was studied.
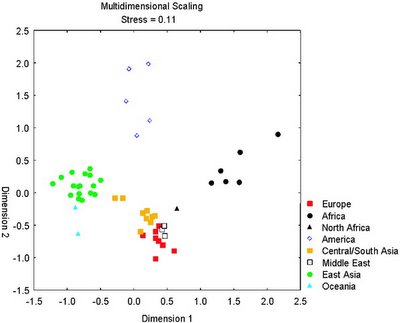
The researchers were able to discern the population origin of these 76 individuals using just 10 SNPs. So, it only takes 10 nucleotides to infer whether someone is Sub-Saharan African, West Eurasian, East Asian, or Native American.
Of course, this ability could be due to overfitting in the small 76-individual sample. So, they tested against the larger 1000+ individual CEPH panel which includes 50+ populations from around the world, and were able to correctly infer the ancestry of all individuals.
Thus, it is demonstrated that a very small set of carefully selected polymorphisms are enough to discover the continental origin of an individual:
Finally, the authors also asked why the very informative SNPs show such high frequency differences in the studied populations:
Thus, although the SNPs from the whole-genome analyses used to identify ancestry-informative markers were noncoding, our data indicate that the significant population differences of the markers with maximum informativeness of ancestry seem to be shaped by positive selection rather than by genetic drift.
Am. J. Hum. Genet. (online early)
Proportioning Whole-Genome Single-Nucleotide–Polymorphism Diversity for the Identification of Geographic Population Structure and Genetic Ancestry
Oscar Lao et al.
The identification of geographic population structure and genetic ancestry on the basis of a minimal set of genetic markers is desirable for a wide range of applications in medical and forensic sciences. However, the absence of sharp discontinuities in the neutral genetic diversity among human populations implies that, in practice, a large number of neutral markers will be required to identify the genetic ancestry of one individual. We showed that it is possible to reduce the amount of markers required for detecting continental population structure to only 10 single-nucleotide polymorphisms (SNPs), by applying a newly developed ascertainment algorithm to Affymetrix GeneChip Mapping 10K SNP array data that we obtained from samples of globally dispersed human individuals (the Y Chromosome Consortium panel). Furthermore, this set of SNPs was able to recover the genetic ancestry of individuals from all four continents represented in the original data set when applied to an independent, much larger, worldwide population data set (Centre d'Etude du Polymorphisme Humain–Human Genome Diversity Project Cell Line Panel). Finally, we provide evidence that the unusual patterns of genetic variation we observed at the respective genomic regions surrounding the five most informative SNPs is in agreement with local positive selection being the explanation for the striking SNP allele-frequency differences we found between continental groups of human populations
Link

No comments:
Post a Comment
Stay on topic. Be polite. Use facts and arguments. Be Brief. Do not post back to back comments in the same thread, unless you absolutely have to. Don't quote excessively. Google before you ask.